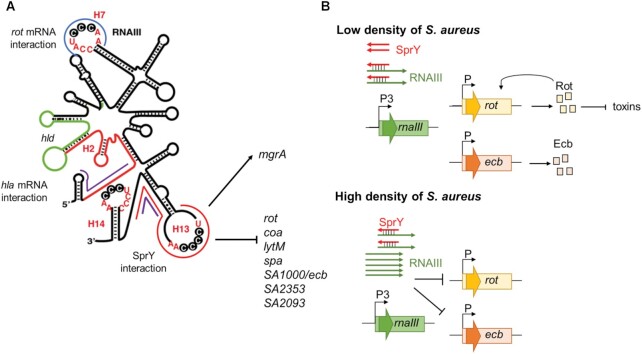
Figure 7.
Schematic view of RNAIII and SprY actions. (A) Schematic view of the secondary structure of RNAIII (12) and base-pairings interaction regions with SprY and other mRNA targets. hla mRNA interacts with RNAIII at 2nd and 3rd hairpins (rnaIII sequence is colored in red). RNAIII encodes the delta-hemolysin (hld, in green). The interaction zone of RNAIII and SprY is indicated in red line and is overlapped with other RNAIII mRNA target listed here. rot mRNA interacts with RNAIII 7th hairpin (in blue line) and RNAIII 13rd stem-loop. mgrA mRNA interacts at two different sites of RNAIII (in purple line). (B) Model for SprY sponge function in RNAIII/ mRNA targets regulation. At low cell density (upper panel), SprY presents in a higher number of copies than RNAIII and bind to all RNAIII molecules produced. This interaction prevents RNAIII from binding to rot and ecb mRNA and favors the translation of Rot and Ecb proteins. At high cell density (bottom panel), RNAIII is produced in higher level than SprY and suppresses the sponge effect of SprY, therefore inhibits the translation of rot and ecb, among other targets.