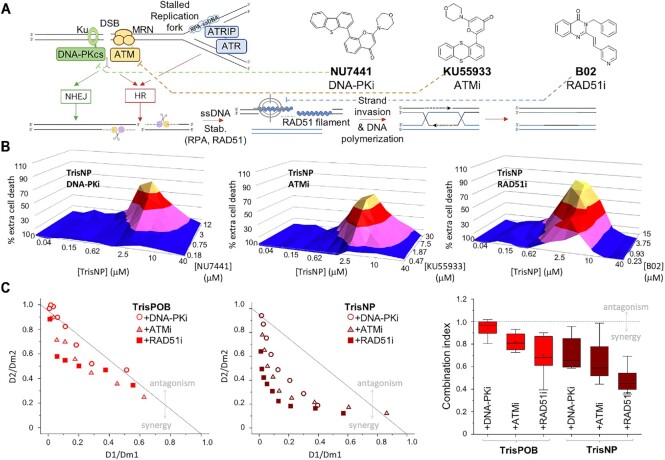
Figure 6.
(A) Double-strand breaks (DSB) and stalled replication forks are repaired by HR and NHEJ, mediated by DNA-PK and ATM kinases and RAD51. Created with BioRender. Chemical structure of DDR inhibitors NU7441 (DNA-PKi), KU55933 (ATMi) and B02 (RAD51i). (B) 3D pyramid plots showing the percentage of extra cell death for the combination of TrisNP (lethal concentration range: 0–40 μM), with DNA-PKi, ATMi, or RAD51i (non-lethal concentration range, see Supplementary Data). Results are the average of three separate experiments each containing technical duplicates. (C) Normalised isobolograms for combination of TrisPOB or TrisNP with DNA-PKi (circles), ATMi (triangles), and RAD51i (squares). Points above the grey horizontal line show antagonism between agents, below the line show synergism. Combination index graph (right) of the same co-treatments. Derived from the same raw data as the pyramid plots in Figure 5B.