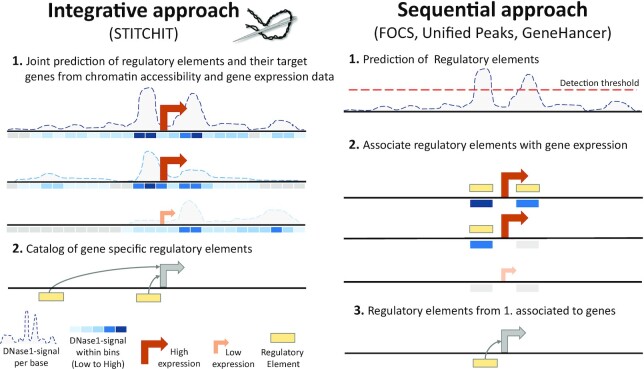
Figure 1.
Comparison of REM inference approaches. (right) Current methods solve the problem of linking REMs to target genes with a sequential approach (e.g. FOCS). Firstly, a catalogue of putative REMs is defined using peak calling. Secondly, signal in REMs in a window around a gene is associated to the expression of the same gene. The final associations are reported (3). (left) STITCHIT combines the step of defining REMs and their association to target genes into a joint prediction problem (1) and generates a catalogue of gene-specific REMs (2.). Thereby REMs that zoom in on the epigenomic region that is associated with gene-expression are generated. This allows to detect more subtle REMs that are missed by sequential approaches that apply strict thresholds on the initial signal.