Figure 2.
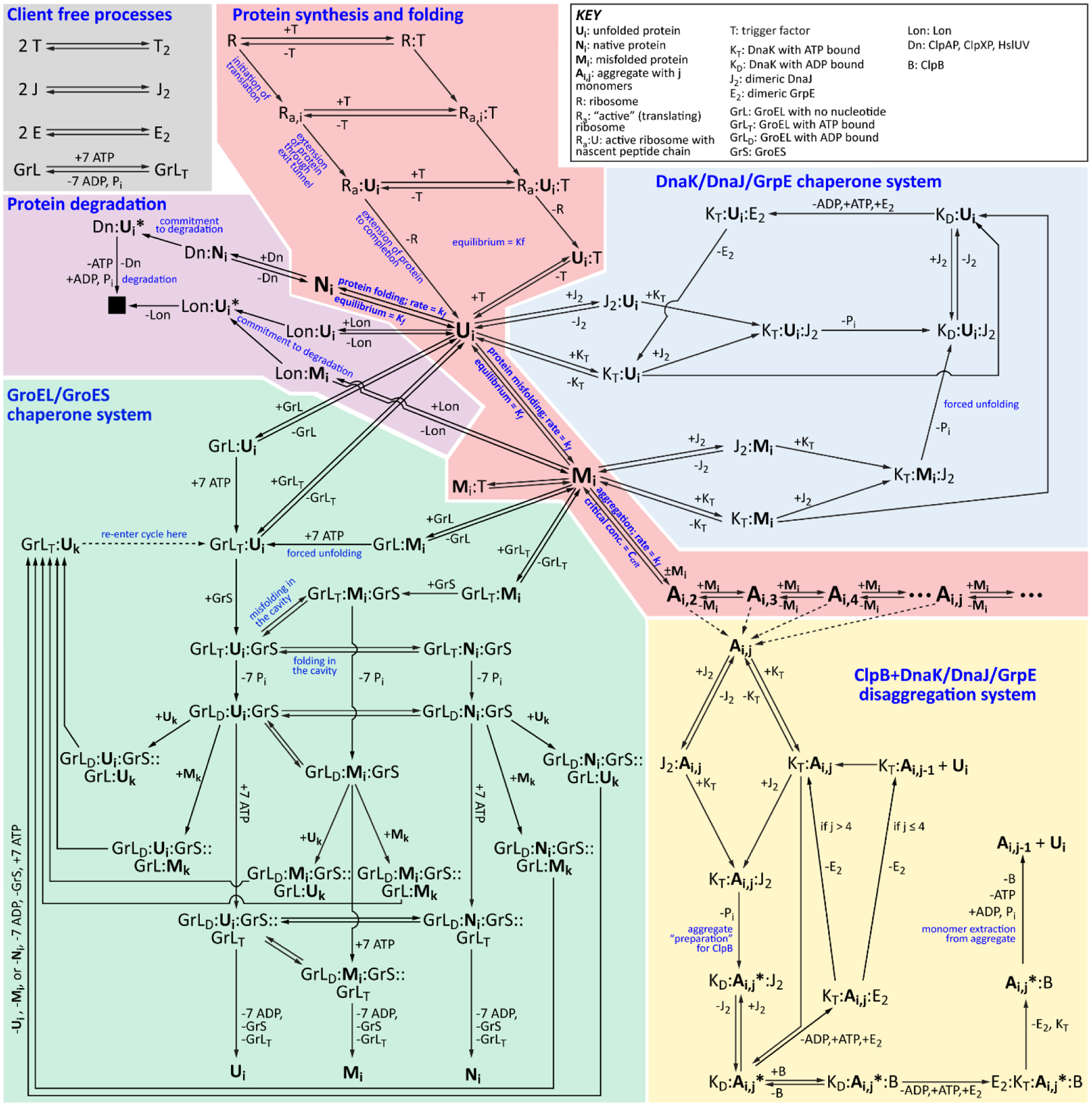
The FoldEco Model for Protein Folding in E. coli. The five subsystems in FoldEco are as follows: protein synthesis and folding (light red), the DnaK/DnaJ/GrpE chaperone system (light blue), the GroEL/GroES chaperone system (light green), the ClpB+DnaK/DnaJ/GrpE disaggregation system (light yellow), and protein degradation (light purple). Also included are several client-free processes, such as DnaJ dimerization (gray). The free unfolded (Ui), misfolded (Mi), native (Ni), and aggregated (Ai,j) states of the client protein are shown in large bold font. Complexes between proteostasis network components and the client protein are denoted by abbreviations for the species in the complex (see key in upper right) separated by colons. When the states of the cis and trans cavities of GroEL are both defined, they are separated by two colons. The subscripts “T” and “D” refer to the ATP- and ADP-bound states of chaperones. Figure and legend reproduced from Powers et al. [110].
