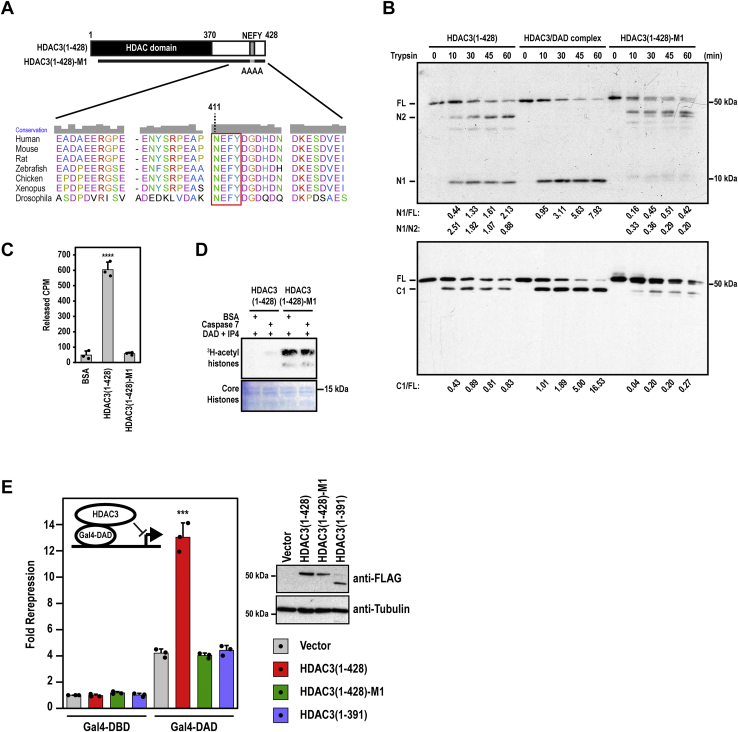
Figure 5.
Sequence-dependent C terminus function and its requirement for DAD activation of HDAC3 in vivo.A, a diagram showing the location of NEFY(411–414), which are mutated to AAAA in HDAC3-M1. Bottom, NEFY(411–414) is conserved across species. B, analysis of the conformational state of HDAC3(1–428)-M1 by partial trypsin digestion assay, along with free WT HDAC3 (left) and the HDAC3–DAD complex (middle). C and D, HDAC activity assays of HDAC3(1–428)-M1 along with the wildtype HDAC3 using liquid scintillation (C) and autoradiographic (D) HDAC assays. In C, the results show mean + standard deviation (n = 3). ∗∗∗∗p ≤ 0.0001 between HDAC3(1–428) and each of the other two samples (two-tailed Student's t test). E, HEK293T cells are transfected with a Gal4-dependent luciferase reporter along with expression plasmids for Gal4–DBD, Gal4–DAD, wildtype or mutant HDAC3 (n = 3). The y-axis denotes fold repression of luciferase activities (Gal4–DBD/Gal4–DAD). The results show mean + standard deviation. ∗∗∗p ≤ 0.001 between HDAC3(1–428) and each of the other samples expressing Gal4–DAD (two-tailed Student's t test). DAD, deacetylase-activation domain; DBD, DNA-binding domain; HDAC, histone deacetylase; IP4, inositol tetraphosphate.