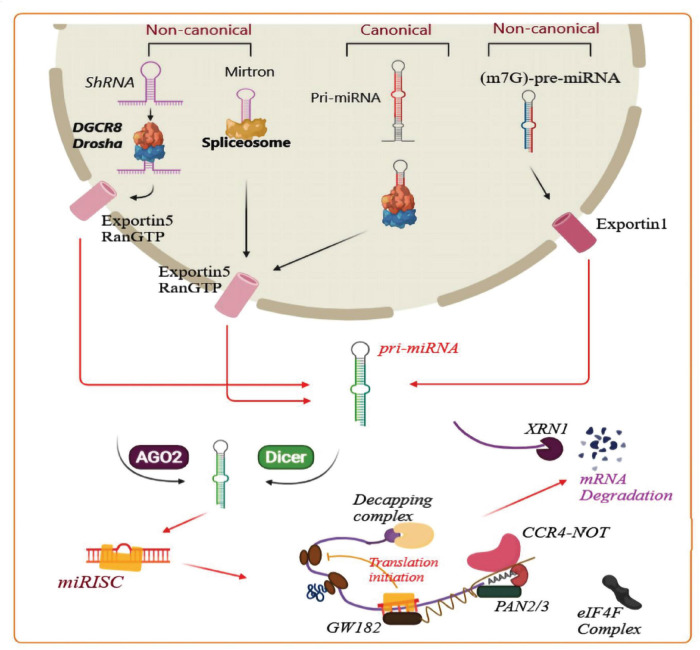
FIGURE 2.
Biogenesis of microRNAs and related mechanisms. A pri-miRNA transcript is the first stage of canonical miRNA biogenesis, which is cleaved by a micro-processor complex (containing Drosha and DGCR8), to create the precursor-miRNA (pre-miRNA). Then, the Exportin5/RanGTP-dependent pathway transports the pre-miRNA to the cytoplasm and processes it to produce a mature miRNA duplex. In the next step, the 5p or 3p strand of the mature miRNA duplex is loaded into a member of the Argonaute (AGO) family of proteins to form a miRNA-induced silencing complex (miRISC). Next, the small hairpin RNA (shRNA) is cleaved in a non-canonical pathway, and Exportin5/RanGTP transports it to the cytoplasm and processes it through an AGO2-dependent, but Dicer-independent cleavage mechanism. 7-Methylguanine capped (m7G)-pre-miRNA and mirtrons depend on Dicer for completing their cytoplasmic maturation, however there are differences in their nucleocytoplasmic shuttling. Exportin5/RanGTP exports mirtrons whereas Exportin1/m7G exports pre-miRNAs. Each pathway results in a functional miRISC complex which can recognize its target mRNA to induce translational suppression via interference with the eIF4F complex. Then, the poly(A)-deadenylases PAN2/3 and CCR4-NOT are recruited by the GW182 family protein attached to Argonaute, and PAN2/3 starts deadenylation while this process is completed by the CCR4-NOT complex, and finally the m7G cap on the target mRNA is removed via the decapping complex. Finally, the decapped mRNA undergoes 5′-3′ degradation through exoribonuclease XRN1 (modified from the results reported in Hayder et al. (2018) study).