Table 1.
AR Binding and Antagonistic Activity of Propanamide Derivatives (13, 19a–n) with Monocyclic B-Ring Modifications
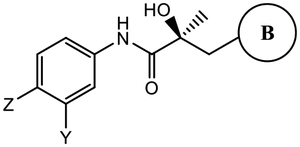
|
|||||
|---|---|---|---|---|---|
| Binding (Ki) / Transactivation (IC50) (μM) | SARD Activity (% degradation) | ||||
| Ki (DHT=1nM)a | IC50b | Full Lengthc (LNCaP) at 1 μM |
Splice Variantc (22RV1) at 10 μM |
||
|
13 (Y=CF3; Z=CN)d |
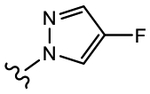
|
>10 | 0.199 | 100 | 100 |
|
19a (Y=CF3; Z=CN) |
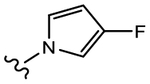
|
0.633 | Partial agoniste | 0 | 0 |
|
19b (Y=CF3; Z=CN) |
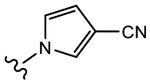
|
0.328 | Partial agonist | 0 | 0 |
|
19c (Y=Cl; Z=CN) |
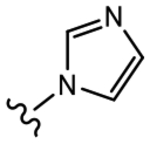
|
>10 | 1.856 | 0 | 30 |
|
19d (Y=CF3; Z=CN) |
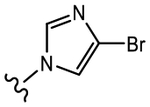
|
0.906 | 0.149 Partial agonist |
0 | 0 |
|
19e (Y=CF3; Z=CN) |
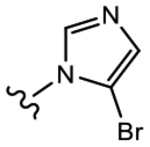
|
>10 | 1.019 | 50 | 70 |
|
19f (Y=CF3; Z=CN) |
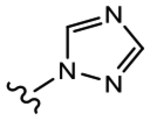
|
>10 | 1.091 | 0 | 0 |
|
19g (Y=CF3; Z=CN) |
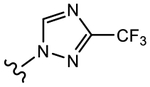
|
>10 | 1.013 | 68 | 100 |
|
19h (Y=Cl; Z=CN) |
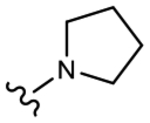
|
>10 | No effect | 0 | 0 |
|
19i (Y=CF3; Z=CN) |
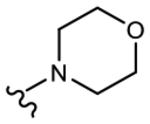
|
1.874 | 1.018 | 52 | 80 |
|
19j (Y=CF3; Z=CN) |
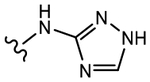
|
>10 | No effect | 0 | N.A.f |
|
19k (Y=CF3; Z=CN) |
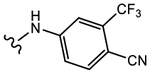
|
>10 | Agonist | 0 | N.A. |
|
19l (Y=CF3; Z=N02) |
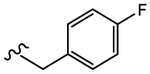
|
N.A. | Agonist | N.A. | N.A. |
|
19m (Y=CF3; Z=CN) |
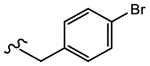
|
0.251 | Agonist | N.A. | N.A. |
|
19n (Y=CF3; Z=CN) |
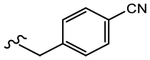
|
0.884 | Agonist | N.A. | N.A. |
AR binding was determined by competitive binding of 1 nM tritiated mibolerone ([3H] MIB) to recombinant LBD of wild-type AR (wtAR). DHT was used in each experiment as a standard agent, and the values are normalized to DHT, with the IC50 of DHT taken as 1 nM.
Inhibition of transactivation was determined by transfecting HEK-293 or COS-7 cells with full-length wtAR, GRE-LUC, and CMV-renilla luciferase for transfection control. Cells were treated 24 h after transfection with a dose–response of compounds (1 pM–10 μM) in the presence of 0.1 nM R1881 (antagonist mode) or in the absence of R1881 (agonist mode). Luciferase assay was performed 24 h after treatment using a dual luciferase (firefly and Renilla) assay kit (Promega, Madison, WI and Goldbio Luciferase kit, St. Louis, MO).
SARD activity was assayed by treating LNCaP or 22RV1 cells for determining FL AR (at 1 μM of antagonist) or AR SV (at 10 μM of antagonist) protein levels, respectively. Cells were maintained in a charcoal-stripped serum-containing medium for 48 h and treated with the indicated doses of antagonist for 24 h in the presence of 0.1 nM R1881 (agonist). Cells were harvested, and Western blot for AR was performed using AR-N20 or PG-21 antibody that is directed toward the NTD of AR and actin (internal control for protein loading). The AR FL and AR SV bands were quantified and normalized to actin bands and represented as percent inhibition from vehicle-treated cells.
The result was reported in the literature in the same assay as described here.40
Transcriptional activation was performed in the same assay in agonist mode.
N.A. means data not available.
