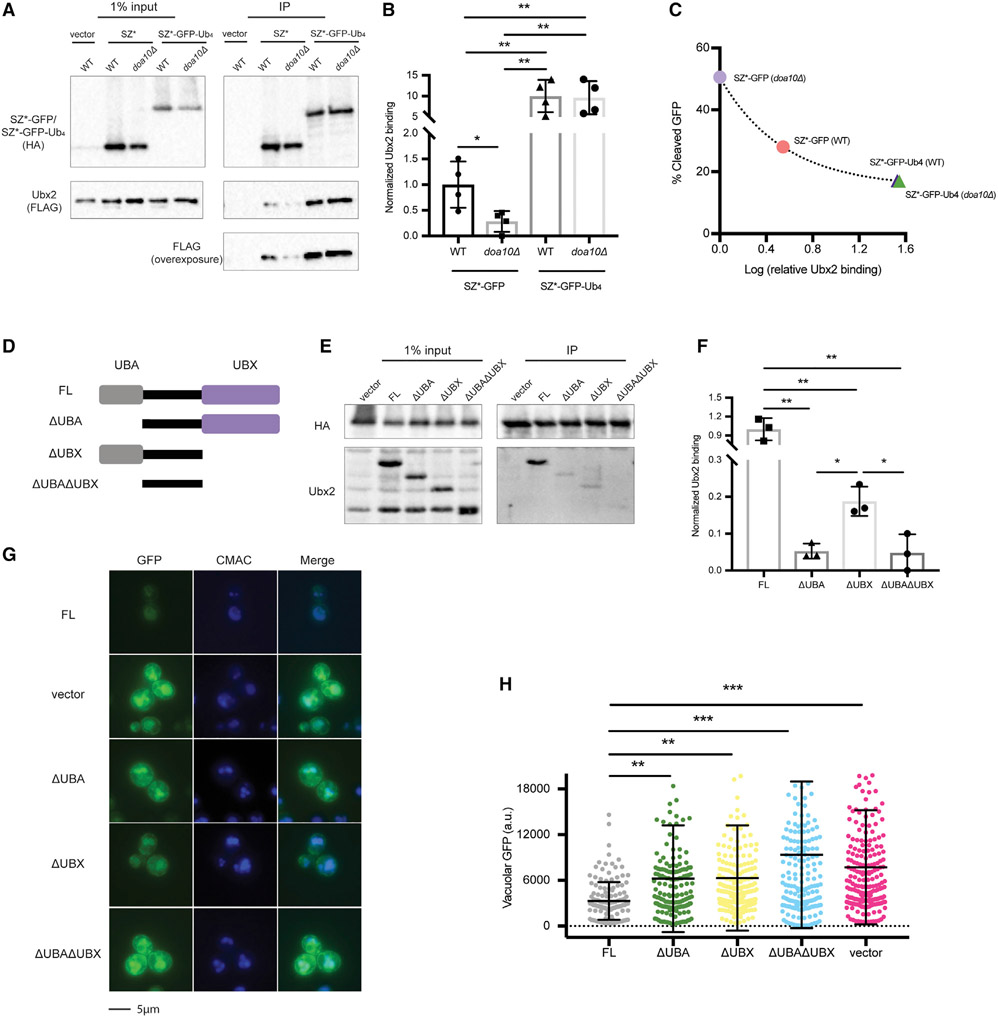
Figure 6. The interaction between SZ* and Ubx2 is enhanced by an appended tetra-ubiquitin moiety.
(A) HA-tagged SZ*-GFP and SZ*-Ub4 were immunoprecipitated from both wild-type and doa10Δ yeast expressing the indicated substrates or containing a vector control. Ubx2 contains a triple FLAG tag. A total of 1% of the input was analyzed to show equal loading.
(B) A bar graph depicts relative Ubx2 binding by SZ*-GFP and SZ*-Ub4 in the indicated yeast strains quantified from (A). Data are the means ± SE of 4 independent experiments; *p < 0.05 and **p < 0.005.
(C) Relative Ubx2 binding was determined by native immunoprecipitation from (A), and percent cleaved GFP at 120 min was determined by pulse-chase from Figure 4F. Data are the means of 4 independent experiments for relative Ubx2 binding and 3–4 independent experiments for percent GFP cleavage.
(D) Schematic of Ubx2 and truncated variants; FL, 1–585 amino acid (aa); ΔUBA, 71–585 aa; ΔUBX, 1–410 aa; ΔUBAΔUBX, 71–410 aa.
(E) SZ*-GFP-Ub4 was immunoprecipitated from ubx2Δ yeast expressing the Ubx2 variants or containing a vector control. After SDS-PAGE, the indicated antibodies were used to detect SZ*-GFP-Ub4 (anti-HA) and the Ubx2 variants. A total of 1% of the input was analyzed to show equal loading.
(F) A bar graph depicts SZ*-Ub4 binding quantified from (E). Data are the means ± SE of 3 independent experiments; *p < 0.05, **p < 0.005.
(G) Live-cell fluorescence imaging of SZ*-GFP in ubx2Δ yeast containing a vector or the indicated Ubx2 variants. CMAC marks the yeast vacuole.
(H) Quantification of GFP intensity in the vacuole in (G). Greater than 100 cells in each strain were counted; **p < 0.005, ***p < 0.0005.