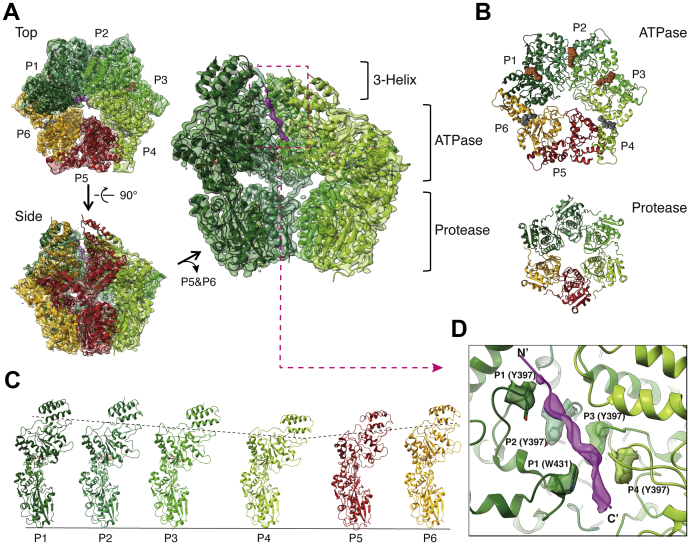
Figure 1.
Overall structure of the substrate-bound MtaLonA complex.A, the ribbon model of Ig2 (magenta) bound MtaLonA fitting into the 3.6-Å cryo-EM map (semitransparent) is presented in the top and side views. The protomers in four substrate-engaged states (P1–P4) are colored in different shades of green; the protomers in two disengaged states (P5 and P6) are colored in brick red and orange, respectively. Protomers P5 and P6 of MtaLonA are removed in the close-up view (right) to show the density of Ig2 (dashed box) inside. B, top views show the rings of the ATPase domains (top) and the protease domains (bottom). ATP-γ-S and ADP are represented by brown and gray spheres, respectively. C, six conformational states (P1–6) of the protomers, arranged in clockwise order in the structure, are shown from left to right; the protomers are aligned using the protease domain as the reference. D, pore-loop 1 (PL1) and 2 (PL2) residues from the protomers P1 to P4 interacting with Ig2 (magenta density) are shown with surrounding density. N′ and C′ denote the N- and C-terminal ends, respectively.