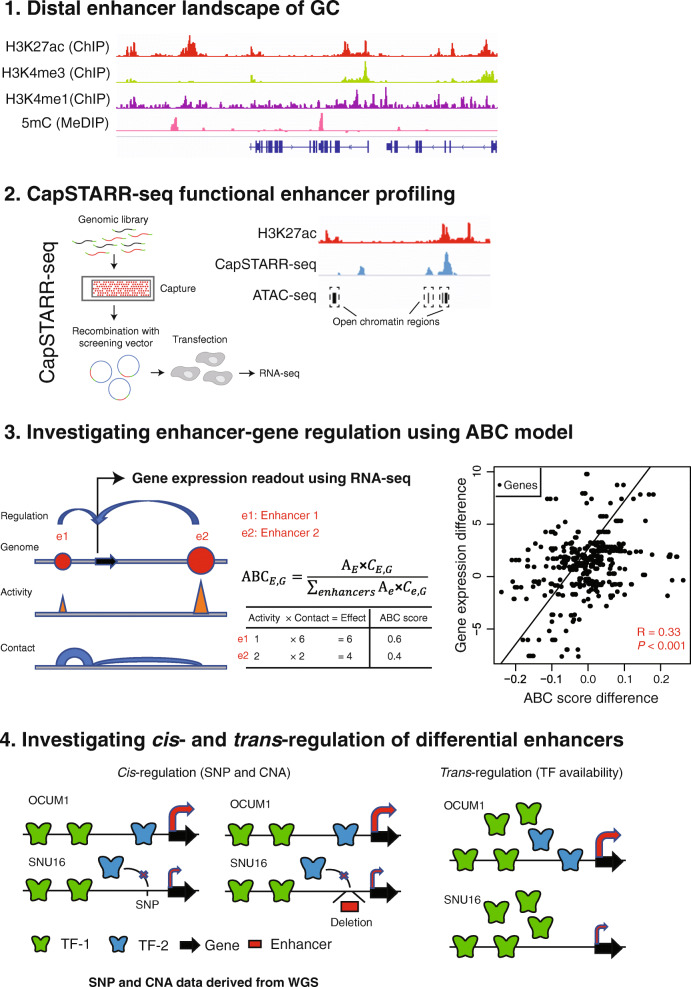
Fig. 1.
Workflow of analyses conducted in this study. The diagram describes the general flow of analyses performed in this study and datasets used. Briefly, distal enhancer landscapes of GC were profiled using H3K27ac ChIP-seq, supported by additional H3K4me3 ChIP-seq, H3K4me1 ChIP-seq, and 5mC MeDIP-seq datasets. CapSTARR-seq profiling was then performed to identify functional enhancers, and integrated with ATAC-seq measuring chromatin accessibility. Enhancer-gene regulation was investigated using the ABC (“Activity-by-contact”) model, where RNA-seq was used as a readout of target gene expression levels. Finally, we examined potential cis- and trans-regulation mechanisms of differential enhancers. For cis-regulation, we derived SNP and CNA (copy number alteration) information using WGS datasets