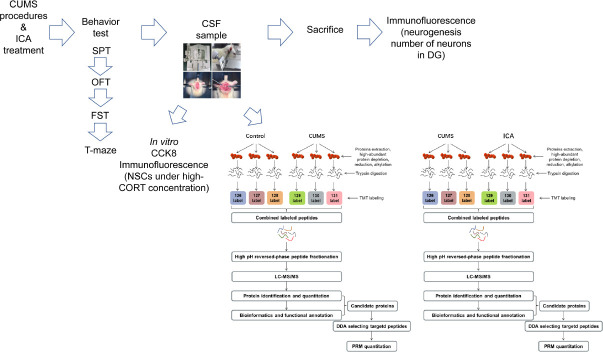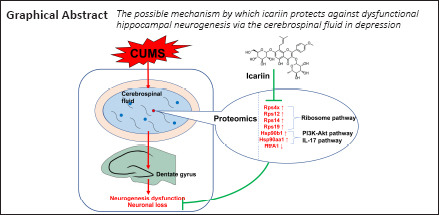
Key Words: cerebrospinal fluid, chronic unpredictable mild stress, depression, dysfunctional hippocampal neurogenesis, hippocampus, icariin, proteomics, ribosome pathway
Abstract
Icariin (ICA) has a significant capacity to protect against depression and hippocampal injury, but it cannot effectively cross the blood-brain barrier and accumulate in the brain. Therefore, the mechanism by which ICA protects against hippocampal injury in depression remains unclear. In this study, we performed proteomics analysis of cerebrospinal fluid to investigate the mechanism by which ICA prevents dysfunctional hippocampal neurogenesis in depression. A rat model of depression was established through exposure to chronic unpredictable mild stress for 6 weeks, after which 120 mg/kg ICA was administered subcutaneously every day. The results showed that ICA alleviated depressive symptoms, learning and memory dysfunction, dysfunctional neurogenesis, and neuronal loss in the dentate gyrus of rats with depression. Neural stem cells from rat embryonic hippocampi were cultured in media containing 20% cerebrospinal fluid from each group of rats and then treated with 100 μM corticosterone. The addition of cerebrospinal fluid from rats treated with ICA largely prevented the corticosterone-mediated inhibition of neuronal proliferation and differentiation. Fifty-two differentially expressed proteins regulated by chronic unpredictable mild stress and ICA were identified through proteomics analysis of cerebrospinal fluid. These proteins were mainly involved in the ribosome, PI3K-Akt signaling, and interleukin-17 signaling pathways. Parallel reaction monitoring mass spectrometry showed that Rps4x, Rps12, Rps14, Rps19, Hsp90b1, and Hsp90aa1 were up-regulated by chronic unpredictable mild stress and down-regulated by ICA. In contrast, HtrA1 was down-regulated by chronic unpredictable mild stress and up-regulated by ICA. These findings suggest that ICA can prevent depression and dysfunctional hippocampal neurogenesis through regulating the expression of certain proteins found in the cerebrospinal fluid. The study was approved by the Experimental Animal Ethics Committee of Guangzhou University of Chinese Medicine of China in March 2017.
Chinese Library Classification No. R453; R749.05; R749.4+1
Introduction
Depression is one of the most common mental disorders, mainly characterized by despair, anhedonia, and even suicide attempts (Bachmann, 2018). As life pressure and social stress increase, the prevalence and morbidity of depression continue to rise (Guo et al., 2020). Depression represents a substantial burden to individuals, their families, and society, making this disorder a worldwide public health concern. Depression is a psychological disease with complex etiology and clinical manifestations, and its pathogenesis has not been fully elucidated.
Hippocampal damage, and specifically dysfunctional hippocampal neurogenesis, is known to play an important role in depression (O’Leary and Cryan, 2014; Park, 2019; Huang et al., 2020). Clinical autopsies have showed reduced hippocampal volume and severe neuronal loss or atrophy in patients with depression (Maller et al., 2007). In addition, preclinical studies have demonstrated reduced numbers of branches, shorter hippocampal neuron dendrites, and impaired neurogenesis in animal models of depression (Danzer, 2012; Duman and Aghajanian, 2012). Therefore, repairing hippocampal damage has become a focus for treating depression (Wu et al., 2013).
Icariin (ICA) is one of the most important bioactive components of Herba epimedii. Recent evidence has confirmed that ICA can effectively alleviate depressive symptoms and protect the hippocampus by reducing neuroinflammation and alleviating hypothalamic-pituitary-adrenal axis dysfunction (Liu et al., 2015, 2019; Wei et al., 2016). However, it cannot effectively cross the blood-brain barrier and is eliminated rapidly from the body, and thus shows very little accumulation in the brain (Chen et al., 2011; Xu et al., 2017). While ICA has a mild direct effect on the brain parenchyma, it is unknown whether it also protects against hippocampal damage in another way.
Cerebrospinal fluid (CSF) is the ultrafiltrate from plasma that is in direct contact with the central nervous system (CNS). CSF surrounds the hippocampus, which is adjacent to the lateral ventricle. Substances in the CSF may directly affect hippocampal structure and function, for example by regulating neurogenesis (Lepko et al., 2019; Planques et al., 2019). CSF plays an important role in molecular exchange and signal transmission in the CNS. Consequently, alteration of CSF components may affect the occurrence and development of CNS diseases (Skipor and Thiery, 2008; Ogawa et al., 2018; Qin et al., 2019). The CSF of patients with depression exhibits altered proteomics, and the differentially expressed proteins are closely linked to CNS damage and dysfunction (Ditzen et al., 2012; Al Shweiki et al., 2017). On the basis of the relationships among CSF components, depression, and hippocampal damage, we hypothesized that ICA influences the brain parenchyma through limbic regions such as the CSF and choroid plexus, thereby exerting anti-depressive effects and protecting against hippocampal damage. To test this this hypothesis in vivo, we used a rat model of depression based on chronic unpredictable mild stress (CUMS). Proteomics analysis was performed to screen for differentially expressed proteins (DEPs) in the CSF co-regulated by CUMS and ICA. We also tested this hypothesis in vitro by simulating stress-induced injury to neural stem cells (NSCs) with a high concentration of corticosterone (CORT) and investigating the effect of ICA-treated CSF on the proliferation and differentiation of these cells. The purpose of this study was to investigate the mechanism through which ICA prevents hippocampal injury in depression.
Materials and Methods
Animals
Since male rodents exposed to CUMS are more stable than females (Franceschelli et al., 2014), 50 male Wistar rats weighing 180–220 g and aged 7–8 weeks were obtained from the Laboratory Animal Center of Southern Medical University, Guangzhou, China (license No. SCXK (Yue) 2016-0041). Rats were housed five per cage and allowed to acclimate to the housing facility for 1 week (23 ± 2°C; 48–60% humidity; 12-hour dark-light cycle; water and food ad libitum). All experimental procedures and protocols were approved by the Experimental Animal Ethics Committee of Guangzhou University of Chinese Medicine of China in March 2017. The experimental procedures were designed in accordance with the United States National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85-23, revised 1996). All efforts were made to minimize animal suffering and to reduce the number of animals used. Before the rats were sacrifices, they were deeply anesthetized by intraperitoneal injection of pentobarbital (30 mg/kg; Sigma, St. Louis, MO, USA).
In vivo drug treatment
After 1 week of acclimatization to the animal facility, all rats were transferred to individual cages to perform the sucrose preference test (SPT). Rats were excluded from further experiments if they exhibited one or more of the following behaviors during the SPT: low sucrose preference (less than 60%), location preference (preferred to drink liquid from a fixed location), drinking too little (drinking neither sucrose solution nor pure water), or excessive drinking (total liquid consumption more than twice the average total liquid consumption of all rats). In total, five rats were excluded due to low sucrose preference (less than 60%).
The remaining 45 rats were randomly divided into three groups: the control (CON, n = 15), CUMS (CUMS, n = 15), and ICA groups (ICA, n = 15). There was no difference in body weight or sucrose preference among these three groups. ICA (purified from Herba epimedii; purity ≥ 98% determined by high performance liquid chromatography) was purchased from Nanjing Dilger Medical Technology Co. Ltd. (Nanjing, China) and dissolved in saline to a concentration of 30 mg/mL. At 16:00 every day during the experimental period, rats in the ICA group received 120 mg/kg ICA intragastrically, and rats in the CON and CUMS groups received the same volume (4 mL/kg) of normal saline intragastrically.
CUMS procedure
Rats in the CON group were housed five per cage and underwent normal feeding without any stressors. Rats in the CUMS and ICA groups were housed individually and subjected to CUMS for 6 weeks. The CUMS procedure used in this study was a modified version of the procedure used in our previous study (Huang et al., 2020). In brief, rats were randomly exposed to 1–2 stressful stimuli once a day for 6 weeks, with no stressors repeated for more than 3 consecutive days (Figure 1). Stressors included white noise (85 dB, 5 hours); thermal swimming (45°C, 5 minutes); stroboscopic illumination (300 flashes/min, 5 hours); soiled cage (10 hours); housing with four other stressed animals (10 hours); cold swimming (4°C, 5 minutes); tail pinching (3 minutes); restraint (12 hours); water deprivation (12 hours); and food deprivation (12 or 24 hours). After 6 weeks of CUMS, behavioral tests were carried out, and CSF and hippocampal samples were taken (Figure 2).
Figure 1.
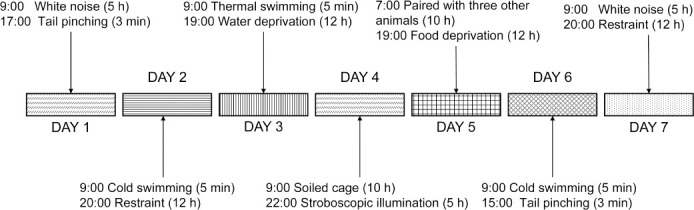
Sample chronic unpredictable mild stress protocol.
One or two arbitrary mild stressors were randomly applied for 6 weeks. None of the stressors were applied for more than 3 consecutive days.
Figure 2.
Experimental procedure.
CCK8: Cell counting kit-8; CORT: corticosterone; CSF: cerebrospinal fluid; CUMS: chronic unpredictable mild stress; DDA: data-dependent acquisition; DG: dentate gyrus; FST: forced swimming test; ICA: icariin; LC-MS/MS: liquid chromatography-tandem mass spectrometry; NSC: neural stem cell; OFT: open field test; PRM: parallel reaction monitoring; SPT: sucrose preference test; TMT: tandem mass tag.
SPT
An SPT was performed to assess anhedonia, as we described previously (Huang et al., 2020). During the SPT, all rats were kept in individual cages. The SPT was divided into four stages: sucrose training for 48 hours, baseline testing for 36 hours, food and water deprivation for 24 hours, and sucrose preference testing for 12 hours. Two bottles of liquid (1% sucrose solution and pure water) were given to each animal at the same time (20:30), and the rats were allowed to drink from both ad libitum for 12 hours (until 8:30 the next day). The volume of each solution that was consumed was then recorded to calculate sucrose preference using the following formula: sucrose preference (%) = sucrose solution consumption/total liquid intake × 100.
Open field test
An open field test (OFT) was performed to assess locomotor activity, as we previously described (Huang et al., 2020). The rats were transferred to the behavioral test room (soundproof dark room) before the OFT and allowed to acclimatize to the environment for 1 hour. Then, each rat was individually placed into the middle of the open-field apparatus (100 cm × 100 cm × 48 cm) and allowed to explore freely for 5 minutes. The total traveling distance was recorded by video-tracking system (Flydy Co., Ltd., Guangzhou, China).
Forced swimming test
Immobile time during a forced swimming test (FST) was used to evaluate despair, as we described previously (Wu et al., 2016). The rats were allowed to acclimatize to the behavioral test room for 1 hour before the test. During the FST, each rats was individually placed in a transparent plexiglas cylinder (height: 100 cm, diameter: 30 cm; Flydy Co., Ltd.) filled with 35 cm of water (25 ± 1°C) and forced to swim for 6 minutes. The amount of time that each rat remained immobile during the last 4 minutes of the test was recorded by three researchers blinded to the experimental design. Rats were considered immobile when they ceased struggling and floated motionless in the water, except for making any movements necessary to keep their heads above the water.
T-maze test
A T-maze test was used to evaluate learning and memory (Yang et al., 2018). The T-maze (Flydy Co., Ltd.) is an elevated maze comprising a start arm (71 cm × 18 cm × 30 cm) and two target arms (46 cm × 18 cm × 30 cm) made of black, non-reflective panels. The test was performed in the dark. Prior to performing the T-maze test, the rats were given only approximately 75% of their usual daily amount of food to ensure that they were hungry before starting the test. The T-maze test was divided into training and testing phases. During the training phase, cheese was placed at the ends of both target arms, with both doors open. The rats were placed in the start arm and allowed to freely explore the maze and consume the cheese. During the testing phase, cheese was placed at the ends of both target arms. The door to one randomly selected target arm, referred to as the goal arm, was closed. The rat was placed in the start arm and allowed to eat the cheese in the open target arm. Once the rat entered the open target arm, the door was closed immediately. When the rat had finished eating, it was removed from the maze. Thirty seconds later, the rat was placed in the start arm again and allowed to freely explore the maze with both doors open. If the rat entered the goal arm and finished eating the cheese, this was scored as one correct run. The testing phase was repeated 10 times a day (each interval was 20 minutes) and lasted for 4 days. Each rat’s accuracy was calculated using the following formula: accuracy (%) = number of correct run/10 × 100 (Yang et al., 2018).
CSF sample preparation
CSF samples were taken 24 hours after the T-maze test. The rats were anesthetized (30 mg/kg, pentobarbital, intraperitoneal injection), and the foramen magnum was exposed. An intravenous infusion needle (0.45#) attached to a 1-mL syringe was used to puncture the cisterna magna and collect the CSF (Zhu et al., 2018), which was then centrifuged at 2000 × g for 15 minutes at 4°C. The supernatant (containing the CSF) was collected and frozen at –80°C. After CSF collection, the rats were sacrificed for brain tissue sampling.
Immunofluorescent analysis of tissue sections
As we described earlier (Huang et al., 2020), 5-bromo-2-deoxyuridine (BrdU, Sigma) was injected intraperitoneally (three injections, 200 mg/kg, 4 hours apart). One week later, the brain tissues (2.7–6.7 mm from the coronal groove) were dissected out and fixed in 4% paraformaldehyde for 24 hours at 4°C, then immersed in 30% sucrose until they sank to the bottom of the tube. The hippocampus was identified on the basis of the distance from the coronary sulcus (2.7–6.7 mm from the coronal sulcus), isolated, and cut into 40 μm–thick slices.
BrdU/doublecortin (DCX) double labeling was used to label the newly formed precursor neurons proliferating and differentiating from NSCs in the dentate gyrus (DG) (Huang et al., 2020). Sections were treated with 2 M HCl for 20 minutes at 37°C, washed thrice in phosphate buffer saline (0.1 M, pH 8.4), blocked in 5% goat serum (Beyotime, Beijing, China; containing 0.03% Triton-X-100) at room temperature for 1 hour, and incubated with rat anti-BrdU (1:200, Cat# ab6326, Abcam, Cambridge, UK) and rabbit anti-DCX (1:200, Cat# ab18723, Abcam) antibodies at 4°C overnight. After washing in Tris-buffered saline with 0.01% Tween-20, the sections were incubated with AlexaFluor® 594 goat anti-rat IgG (1:500, Cat# ab150160, Abcam) and AlexaFluor® 488 goat anti-rabbit IgG (1:500, Cat# ab150077, Abcam) at 37°C for 2 hours. After washing and 4′,6-diamidino-2-phenylindole staining, images were captured by a laser confocal microscope (LSM800, ZEISS, Oberkochen, Germany). The number of BrdU/DCX double-positive cells was recorded.
NeuN was used to label and count mature neurons in the DG (Huang et al., 2020). The sections were thawed, subjected to membrane rupture using Triton-X-100, blocked with goat serum, and incubated with a rabbit anti-NeuN antibody (1:1000, Cat# ab177487, Abcam) overnight at 4°C. After washing in Tris-buffered saline with 0.01% Tween-20, the sections were incubated with AlexaFluor® 488 goat anti-rabbit IgG (1:500, Cat# ab150077, Abcam) at 37°C for 2 hours. After washing and 4′,6-diamidino-2-phenylindole staining, images were captured by a laser confocal microscope. The proportion of NeuN-positive cells (%) was calculated as follows: NeuN-positive cell number/total number of nuclei × 100.
Primary hippocampal NSC culturing conditions
Embryos were removed from etherized Wistar rats (obtained from the Laboratory Animal Center of Southern Medical University, Guangzhou, China) on embryonic days 16–18 under sterile conditions. The bilateral hippocampi were dissected out, cut into 1 mm × 1 mm × 1 mm pieces, and placed in ice-cold sterile phosphate buffer saline. The solution was then pipetted gently up and down with a Pasteur pipette to dissociate the tissues, filtered through a 200-mesh cell sieve to obtain a single-cell suspension, and centrifuged at 260 × g for 5 minutes. The supernatant was discarded, and the cells were re-suspended with NSC medium (Dulbecco’s modified Eagle media/Nutrient mixture F-12 (Gibco, Grand Island, NY, USA) supplemented with 20 ng/L epidermal growth factor (Gibco), 20 ng/L basic fibroblast growth factor (Gibco), 2% B27 (Gibco) and 1% penicillin/streptomycin (Gibco)). Cells were seeded into 60-mm culture dishes at a density of 2 × 105 cells/mL and incubated at 37°C in a 5% (v/v) CO2 incubator. Half of the culture medium was replaced with fresh medium every 2–3 days. Seven days later, the newly formed neurospheres were digested with AccutaseTM (Gibco) to dissociate them for passaging.
Cell counting kit-8 test
Passage 1 NSCs were collected and divided into five groups: CON (cultured with NSC medium (Dulbecco’s modified Eagle media/Nutrient mixture F-12 containing 20 ng/L epidermal growth factor, 20 ng/L basic fibroblast growth factor, 2% B27 and 1% penicillin/streptomycin)), high-concentration CORT (cultured with NSC medium containing 100 μM CORT (Millipore, Billerica, CA, USA)), CON-CSF (cultured with NSC medium containing 20% CSF harvested from CON rats (CON-CSF) +100 μM CORT), CUMS-CSF (cultured with NSC medium containing 20% CSF harvested from CUMS rats (CUMS-CSF) + 100 μM CORT), and ICA-CSF (cultured with NSC medium containing 20% CSF harvested from ICA rats (ICA-CSF) + 100 μM CORT).
Cell viability was detected using a cell counting kit-8 (CCK8). As we described previously (Wu et al., 2013), P1 neutrospheres were digested into single cells by AccutaseTM (Gibco), re-suspended with NSC medium, and seeded in 96-well plates at a density of 4 × 104 cells/well. CSF and CORT were added as appropriate, for a final volume of 100 μL per well. After 72 hours, 10 μL of CCK8 reagent (Dojindo, Kumamoto, Japan) was added to each well, and the cells were incubated for another 2 hours. The NSCs were then dispersed into a single-cell solution by enzymatic digestion, re-suspended with NSC culture medium, and seeded into 96-well plates at a density of 4 × 104 cells per well. The optical density (OD) was measured at 450 nm with a microplate reader (Bio-Rad, Hercules, CA, USA). The cell viability was calculated as following: cell viability (%) = (ODexperimental– OD blank) / (ODCON – ODblank) × 100.
Immunofluorescent analysis of hippocampal NSCs
We previously used media containing 10% fetal bovine serum (FBS) as a differentiation medium (Wu et al., 2013). Given that CSF plays a role in promoting NSC differentiation (Lepko et al., 2019; Planques et al., 2019), NeuralBasal (Gibco) containing 2% FBS (Gibco) was used as a differentiation culture medium in this study. Passage 1 NSCs were collected and divided into seven groups: control 1 (cultured with NeuralBasal containing 10% FBS), control 2 (cultured with NeuralBasal containing 2% FBS), control 3 (cultured with NeuralBasal containing 2% FBS + 20% normal CSF), high-concentration CORT (cultured with NeuralBasal containing 2% FBS + 100 μM CORT), CON-CSF (cultured with NeuralBasal containing 2% FBS + 100 μM CORT + 20% CSF from CON rats), CUMS-CSF (cultured with NeuralBasal containing 2% FBS + 100 μM CORT + 20% CSF from CUMS rats), and ICA-CSF (cultured with NeuralBasal containing 2% FBS + 100 μM CORT + 20% CSF from ICA rats).
As we previously described (Wu et al., 2013), NSCs were dispersed into single cells using AccutaseTM, re-suspended in the differentiation culture medium (NeuralBasal containing 2% FBS), and seeded into 15-mm confocal dishes (5000 cells/dish). After 48 hours, CORT and CSF were added to the appropriate groups, and the cells were incubated for another 48 hours, after which 10 μM BrdU was added to each dish. After 48 hours, the cells were fixed with 4% paraformaldehyde and labeled with BrdU/DCX. The number of BrdU/DCX double-positive cells and the number of nuclei were counted using the Photoshop (Adobe Photoshop Inc., San Jose, CA, USA) counting tool. The proportion of BrdU/DCX double-positive cells (%) was calculated as follows: number of BrdU/DCX double-positive cells/total number of nuclei × 100.
Tandem mass tag analysis of CSF proteomics
Total protein was extracted from CSF. After quantification and separation, 30 μL of protein from each sample was subjected to proteolysis. Tandem mass tag (TMT) labeling was carried out according to the instructions provided by the manufacturer of the TMT labeling kit (Thermo, Waltham, MA, USA). A high-pH RP spin column was used for grading. The samples were separated by chromatography (Easy nLC; Thermo) and analyzed by mass spectrometry (Thermo). The raw data were identified and quantitatively analyzed using Mascot2.2 (Matrix Science, Boston, MA, USA) and Proteome Discoverer1.4 (Thermo). Proteins with a fold change more than 1.2 or less than 0.83, as well as a statistical P-value < 0.05 between two groups, were selected as DEPs.
Bioinformatic analysis
Gene Ontology (GO) mapping and protein annotation were conducted using Blast2GO (https://www.blast2go.com). Kyoto Encyclopedia of Genes and Genomes (KEGG) annotation was performed using KAAS (KEGG Automatic Annotation Server, http://www.genome.jp/kegg/kaas/). Enrichment analysis was performed by Fisher’s exact test, with P-values < 0.05 and a false discovery rate < 0.05.
Quantitative analysis of target proteins by parallel reaction monitoring
Total protein was extracted from the CSF samples, hydrolyzed, and separated by high performance liquid chromatography (Thermo). The separated peptides were analyzed by parallel reaction monitoring (PRM) mass spectrometry (Thermo). The PRM test was repeated three times. Finally, Skyline3.7.0 software (http://proteome.gs.washington.edu/software/skyline) was used to analyze the data from the original PRM files and to quantify target proteins and peptides.
Statistical analysis
The data were statistically analyzed using SPSS 22.0 software (IBM, Armonk, IL, USA). All data are expressed as the mean ± standard error of mean (SEM). All data in each group were consistent with a normal distribution (Shapiro-Wilk test). One-way analysis of variance was used for comparisons between three or more groups with homogeneous variance, and Welch’s test was used to compare groups with heterogeneous variance. For pairwise comparisons, the least significant difference test was used to assess results with homogeneous variance (FST results, the number of BrdU/DCX double positive cells in the DG, proportion of NeuN-positive cells, CCK8 results, ratio of BrdU/DCX double-positive cells in vitro), and the Games-Howell method was used to assess results with heterogeneous variance (SPT results, OFT results, the number of BrdU-positive cells in vitro). T-maze accuracy was analyzed by repeated-measures analysis of variance. The Greenhouse-Geisser correction was used when the assumption of sphericity was not met. The accuracy between groups at each individual time point was compared by multivariate analysis of variance (T-maze accuracy). P-values < 0.05 were considered statistically significant.
Results
ICA reverses depression-like behaviors in CUMS rats
SPT
As shown in Figure 3A, compared with the CON group, the sucrose preference of the CUMS group was significantly decreased (n = 15, F(2, 42) = 27.476, P < 0.01). Compared with the CUMS group, the sucrose preference of the ICA group was significantly increased (n = 15, F(2, 42) = 27.476, P < 0.01), indicating that ICA can alleviate anhedonia in CUMS rats.
Figure 3.
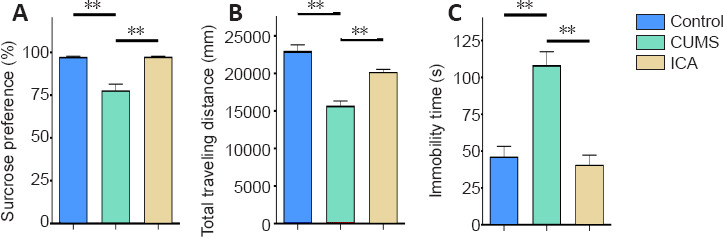
Behavioral effects of ICA in a rat model of depression.
(A) Sucrose preference in the sucrose preference test. Sucrose preference (%) = sucrose solution consumption/total liquid intake × 100. (B) Total traveling distance in 5 minutes in the open field test. (C) Immobile time in the forced swim test. Data are expressed as the mean ± SEM (n = 15 per group), and were analyzed by one-way analysis of variance followed by the least significant difference (C) or Games-Howell (A and B) post hoc test. **P < 0.01. CUMS: Chronic unpredictable mild stress; ICA: icariin.
OFT
As shown in Figure 3B, compared with the CON group, CUMS significantly reduced the total traveling distance (n = 15, F(2, 42) = 34.641, P < 0.01) in the OFT. Compared with the CUMS group, ICA significantly elevated the total traveling distance (n = 15, F(2, 42) = 34.641, P < 0.01), suggesting that ICA has improves locomotor activity in CUMS rats.
FST
As shown in Figure 3C, the immobile time in rats with CUMS was significantly increased compared with the CON group (n = 15, F(2, 42) = 24.394, P < 0.01). ICA significantly decreased the immobile time compared with the CUMS group (n = 15, F(2, 42) = 24.394, P < 0.01). This suggests that ICA treatment can alleviate despair in CUMS rats.
ICA alleviates hippocampal damage in CUMS rats
T-maze accuracy
As shown in Figure 4A, compared with the CON group, there was no difference in the T-maze accuracy of the CUMS group on day 1 (n = 7–10, F(2, 21) = 1.197, P > 0.05), and a significant reduction in accuracy on days 2–4 (n = 7–10, day 2: F(2, 21) = 8.859, P < 0.01; day 3: F(2, 21) = 4.612, P < 0.01; day 4: F(2, 21) = 5.998, P < 0.01). Compared with the CUMS group, the T-maze accuracy of the ICA group on days 2–4 was significantly improved (n = 7–10, day 2: F(2, 21) = 8.859, P < 0.05; day 3: F(2, 21) = 4.612, P < 0.01; day 4: F(2, 21) = 5.998, P < 0.05). This suggests that ICA treatment can improve learning and memory in CUMS rats.
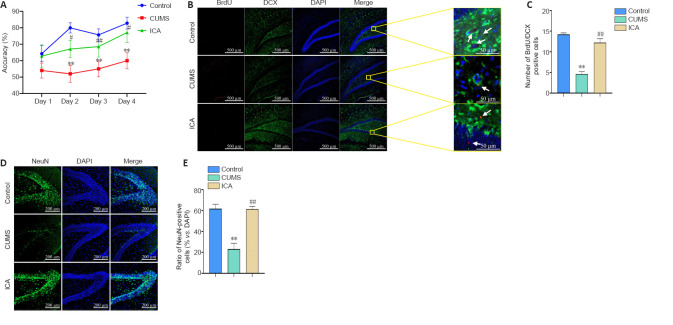
Figure 4.
ICA effects on hippocampal damage in a rat model of depression.
(A) Accuracy in the T-maze test (n = 7–10 per group). Accuracy (%) = correct times/10 × 100. (B) BrdU/DCX double-positive cells (arrows) in the dentate gyrus. ICA significantly increased the number of BrdU/DCX double-positive cells in the dentate gyrus compared with the CUMS group. BrdU: red, AlexaFluor®594; DCX: green, AlexaFluor®488; DAPI: blue. Scale bars: 500 μm; original magnification, 10×; 50 μm in enlarged images. (C) Numbers of BrdU/DCX double-positive cells in the dentate gyrus (n = 5 per group). (D) NeuN-positive cells in the dentate gyrus. ICA significantly increased the proportion of neurons compared with the CUMS group. NeuN: green, AlexaFluor®488; DAPI: blue. Scale bars: 200 μm; original magnification, 20×. (E) Ratio of NeuN-positive cells to total cells in the dentate gyrus (n = 4–5 per group). Data are expressed as the mean ± SEM, and were analyzed by one-way analysis of variance followed by the least significant difference (C, E). **P < 0.01, vs. control group; ##P < 0.01, vs. CUMS group. BrdU: 5-Bromo-2-deoxyuridine; CUMS: chronic unpredictable mild stress; DAPI: 4’,6-diamidino-2-phenylindole; DCX: doublecortin; ICA: icariin.
Number of BrdU/DCX double-positive cells in the DG
As shown in Figure 4B and C, compared with the CON group, the number of BrdU/DCX double-positive cells in the DG in the CUMS group was significantly reduced (n = 5, F(2, 12) = 53.44, P < 0.01). Compared with the CUMS group, treatment with ICA significantly increased the number of BrdU/DCX double-positive cells in the DG (n = 5, F(2, 12) = 53.44, P < 0.01). This indicates that ICA treatment can alleviate CUMS-induced dysfunctional neurogenesis in the DG.
Proportion of NeuN-positive cells in the DG
As shown in Figure 4D and E, compared with the CON group, the proportion of NeuN-positive cells in the DG of the CUMS group was significantly reduced (n = 4–5, F(2, 11) = 24.54, P < 0.01). Compared with the CUMS group, treatment with ICA resulted in a significantly increased proportion of the proportion of NeuN-positive cells (n = 4–5, F(2, 11) = 24.54, P < 0.01). It suggests that ICA treatment can resist neuronal reduction in the DG.
Effect of ICA CSF on the proliferation and differentiation of NSCs exposed to a high concentration of CORT
Proliferation of NSCs exposed to a high concentration of CORT
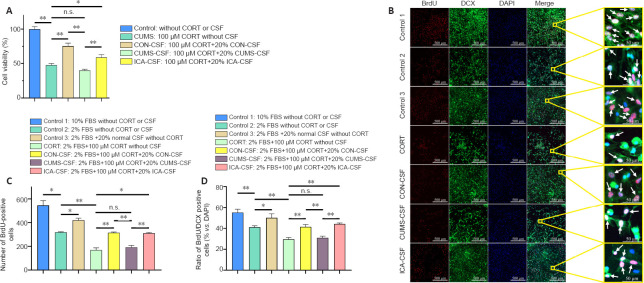
As shown in Figure 5A, compared with the CON group, the viability of NSCs in the CORT group was significantly reduced (n = 10, F(4, 45) = 53.41, P < 0.01). Compared with the CORT group, the NSC viability in the CON-CSF (n = 10, F(4, 45) = 53.41, P < 0.01) and ICA-CSF groups (n = 10, F(4, 45) = 53.41, P < 0.05) was significantly increased. There was no significant difference in NSC viability between the CORT and CUMS-CSF groups (n = 10, F(4, 45) = 53.41, P = 0.105), suggesting that CON-CSF and ICA-CSF promote NSC proliferation in the presence of high concentrations of CORT. In contrast, CUMS-CSF had no significant effect. Compared with the CUMS-CSF group, NSC viability in the ICA-CSF group was significantly increased.
Figure 5.
The effect of CSF on primary hippocampal NSC proliferation and differentiation into neurons exposed to a high concentration of CORT.
(A) Cell viability as detected by Cell Counting Kit-8 (n = 10 per group). Cell viability (%) = [ODexperimental– ODblank]/[ODcontrol– ODblank] × 100. (B) BrdU/DCX double-positive cells (arrows). Normal CSF promoted NSC proliferation and differentiation. Compared with the CUMS-CSF group, the ratio of BrdU/DCX double-positive cells to total cells in the ICA-CSF group was significantly increased. BrdU: red, AlexaFluor®594; DCX: green, AlexaFluor®488; DAPI: blue. Scale bars: 500 μm; original magnification, 10×; 50 μm in enlarged images. (C) The number of BrdU-positive cells (n = 4–7 per group). (D) Ratio of BrdU/DCX double-positive cells to total cells (n = 4–7 per group). Data are expressed as the mean ± SEM, and were analyzed by one-way analysis of variance followed by the least significant difference (A, D) or Games-Howell (C) post hoc test. *P < 0.05, **P < 0.01. BrdU: 5-Bromo-2-deoxyuridine; CON: control; CORT: corticosterone; CSF: cerebrospinal fluid; CUMS: chronic unpredictable mild stress; DAPI: 4′,6-diamidino-2-phenylindole; DCX: doublecortin; FBS: fetal bovine serum; ICA: cerebrospinal fluid; n.s.: not significant; NSC: neural stem cell.
Differentiation of NSCs exposed to a high concentration of CORT
As shown in Figure 5B–D, compared with the control 1 group (10% FBS), the number of BrdU- positive cells (n = 4–7, F(6, 28) = 45.24, P < 0.05) and the proportion of BrdU/DCX double-positive cells (n = 4–6, F(6, 29) = 15.34, P < 0.01) in the control 2 group (2% FBS) were significantly reduced, while there was no significant difference compared with the control 3 group (2% FBS + 20% normal CSF) (n = 4–7, F(6, 28) = 45.24, P = 0.201; n = 4–6, F(6, 29) = 15.34, P = 0.148). Compared with the control 2 group, the number of BrdU-positive cells (n = 4–6, F(6, 28) = 45.24, P < 0.05) and the ratio of BrdU/DCX double-positive cells (n = 4–6, F(6, 29) = 15.34, P < 0.05) in the control 3 group were significantly increased. This suggests that normal CSF promotes NSC proliferation and differentiation.
Compared with the CORT group, the number of BrdU-positive cells (n = 4–7, F(6, 28) = 45.24, P < 0.01) and the ratio of BrdU/DCX double-positive cells (n = 4–6, F(6, 29) = 15.34, P < 0.01) in the CON-CSF group were significantly increased, but there was no significant difference compared with the CUMS-CSF group (n = 4–7, F(6, 28) = 45.24, P = 0.942; n = 4–6, F(6, 29) = 15.34, P = 0.681).
Compared with the CON-CSF group, the number of BrdU-positive cells (n = 4–7, F(6, 28) = 45.24, P < 0.01) and the ratio of BrdU/DCX double-positive cells (n = 4–6, F(6, 29) = 15.34, P < 0.01) in the CUMS-CSF group were significantly reduced, while there was no significant change compared with the ICA-CSF group (n = 4–7, F(6, 28) = 45.24, P = 1.000; n = 4–6, F(6, 29) = 15.34, P = 0.409). Compared with the CUMS-CSF group, the number of BrdU-positive cells (n = 4–7, F(6, 28) = 45.24, P < 0.01) and the ratio of BrdU/DCX double-positive cells (n = 4–6, F(6, 29) = 15.34, P < 0.01) in the ICA-CSF group were significantly increased.
Treatment with ICA results in DEPs in the CSF of rats subjected to CUMS
DEPs in the CSF of rats subjected to CUMS
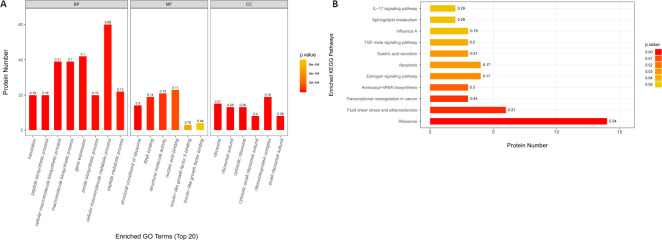
The TMT proteomics screen identified 1935 DEPs between the CON and CUMS groups. One hundred DEPs were selected that exhibited a fold change (CUMS/CON or ICA/CUMS) of ≥ 1.2 or ≤ 0.83 and a P-value of ≤ 0.05 (Additional Table 1). Among these DEPs, 66 were up-regulated compared with the CON group, and 34 were down-regulated (Additional Table 1). GO showed that these proteins are involved in binding, catalytic activity, structural molecule activity, molecular function regulation, and transcriptional regulation. The biological processes they mainly participated in are cellular processes, metabolic processes, biological regulation, regulation of biological processes, and stimulus response. KEGG pathway annotation indicated that these 100 DEPs were mostly involved in ribosomes, fluid shear stress and atherosclerosis, transcriptional misregulation in cancer, aminoacyl-transfer RNA biosynthesis, and the estrogen signaling pathway (Figure 6).
Additional Table 1.
Differentially expressed proteins of cerebrospinal fluid regulated by chronic unpredictable mild stress (CUMS)
| Accession | Protein Name | Gene Name | CUMS/CON |
|---|---|---|---|
| CUMS/CON<1.2 | |||
| ENSRNOP00000002356 | Xyloside xylosyltransferase 1 | Xxyltl | 0.747397003 |
| ENSRNOP00000002394 | Chordin | Chrd | 0.751508465 |
| ENSRN0P00000004313 | Serpin family F member 1 | Serpinfl | 0.813794136 |
| ENSRN0P00000004386 | Myocilin | Myoc | 0.807860809 |
| ENSRN0P00000004764 | EGF containing fibulin extracellular matrix protein 1 | Efempl | 0.824967476 |
| ENSRN0P00000005863 | Lipin 1 | Lpinl | 0.537540772 |
| ENSRN0P00000006070 | Decorin | Dcn | 0.78396755 |
| ENSRN0P00000012286 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 | B3gnt2 | 0.742646316 |
| ENSRN0P00000012293 | Alpha-2u globulin PGCL3 | LOC259244 | 0.782606722 |
| ENSRN0P00000013896 | Serine (or cysteine) proteinase inhibitor, clade A, member 3C | Serpina3c | 0.808979674 |
| ENSRN0P00000014807 | Insulin-like growth factor binding protein 6 | Igfbp6 | 0.778601538 |
| ENSRN0P00000016389 | Transforming growth factor, beta induced | Tgfbi | 0.801979285 |
| ENSRN0P00000017782 | Arylsulfatase A | Arsa | 0.827573422 |
| ENSRN0P00000018359 | Bone morphogenetic protein 6 | Bmp6 | 0.808422469 |
| ENSRN0P00000022480 | Scavenger receptor cysteine rich family member with 5 domains | Ssc5d | 0.723315795 |
| ENSRN0P00000022679 | Matrix metallopeptidase 2 | Mmp2 | 0.811117501 |
| ENSRN0P00000023078 | Protein phosphatase 5, catalytic subunit | Ppp5c | 0.718673731 |
| ENSRN0P00000023530 | Insulin-like growth factor binding protein 5 | Igfbp5 | 0.690951779 |
| ENSRN0P00000025857 | Gelsolin | Gsn | 0.797663166 |
| ENSRN0P00000026583 | Lecithin cholesterol acyltransferase | Lcat | 0.808971821 |
| ENSRN0P00000027860 | HtrA serine peptidase 1 | Htra1 | 0.801966422 |
| ENSRN0P00000033206 | Chloride intracellular channel 6 | Clic6 | 0.246294135 |
| ENSRN0P00000039770 | ADP-ribosyltransferase 3 | Art3 | 0.821875026 |
| ENSRN0P00000047793 | Ret proto-oncogene | Ret | 0.815240431 |
| ENSRN0P00000059194 | Angiopoietin-like 1 | Angptl1 | 0.772500228 |
| ENSRN0P00000068985 | Pappalysin 2 | Pappa2 | 0.733119556 |
| ENSRN0P00000069539 | MIA SH3 domain ER export factor 3 | AABR07021988.1 | 0.710910413 |
| ENSRN0P00000070498 | Carboxypeptidase D | Cpd | 0.75711174 |
| ENSRN0P00000071077 | Collagen type II alpha 1 chain | Col2a1 | 0.544458879 |
| ENSRN0P00000073137 | Insulin-like growth factor binding protein 3 | Igfbp3 | 0.77596736 |
| ENSRN0P00000073724 | Periostin | Postn | 0.786532673 |
| ENSRN0P00000073746 | Cerebellin 3 precursor | Cbln3 | 0.64350437 |
| ENSRN0P00000074387 | Complement C7 | C7 | 0.808958733 |
| ENSRN0P00000075712 | Glypican 3 | Gpc3 | 0.732900873 |
| CUMS/CON>0.8 | |||
| ENSRN0P00000001397 | Transmembrane p24 trafficking protein 2 | Tmed2 | 2.111152161 |
| ENSRN0P00000001518 | Ribosomal protein lateral stalk subunit P0 | Rplp0 | 1.435723382 |
| ENSRN0P00000001593 | Cystatin B | Cstb | 1.634948663 |
| ENSRN0P00000004278 | Ribosomal protein S4, X-linked | Rps4x | 2.82744659 |
| ENSRN0P00000004867 | Small ubiquitin-like modifier 2 | Sumo2 | 1.33984139 |
| ENSRN0P00000008509 | Eukaryotic translation initiation factor 1A, X-linked | Eif1ax | 1.518261651 |
| ENSRN0P00000009028 | Spectrin, beta, erythrocytic | Sptb | 1.227594859 |
| ENSRN0P00000009249 | Proteasome 26S subunit, non-ATPase 6 | Psmd6 | 1.816104855 |
| ENSRN0P00000009556 | Heat shock protein HSP 90-alpha | Hsp90aa1 | 1.368402759 |
| ENSRN0P00000009649 | Proteasome 26S subunit, ATPase 6 | Psmc6 | 1.392119767 |
| ENSRN0P00000010674 | Tyrosyl-tRNA synthetase | Yars | 1.797768969 |
| ENSRN0P00000012726 | Angiopoietin like 7 | Angptl7 | 1.34633353 |
| ENSRN0P00000013375 | Eukaryotic translation initiation factor 2 subunit alpha | Eif2s1 | 2.148860137 |
| ENSRN0P00000015076 | ATPase Na+/K+ transporting subunit beta 2 | Atp1b2 | 1.343898165 |
| ENSRN0P00000015598 | RAB11 a, member RAS oncogene family | Rab11a | 1.368097606 |
| ENSRN0P00000015612 | S100 calcium binding protein A6 | S100a6 | 1.653463759 |
| ENSRN0P00000016036 | LIF receptor alpha | Lifr | 1.210574611 |
| ENSRN0P00000017234 | Heparin binding growth factor | Hdgf | 1.295620414 |
| ENSRN0P00000019162 | Ribosomal protein L35 | Rpl35 | 1.632626406 |
| ENSRN0P00000019247 | Ribosomal protein L27a | Rpl27a | 1.485330137 |
| ENSRN0P00000021048 | Myosin light chain 12A | Myl12a | 2.016783674 |
| ENSRN0P00000022184 | Ribosomal protein S12 | Rps12 | 3.913067297 |
| ENSRN0P00000022603 | Calmodulin 1 | Calm1 | 1.570272851 |
| ENSRN0P00000023935 | Ribosomal protein S3 | Rps3 | 1.925279888 |
| ENSRN0P00000024430 | Vimentin | Vim | 1.374326752 |
| ENSRN0P00000025217 | Ribosomal protein L17 | Rpl17 | 2.072616181 |
| ENSRN0P00000025881 | N-deacetylase and N-sulfotransferase 1 | Ndst1 | 1.253585254 |
| ENSRN0P00000026528 | Ribosomal protein S5 | Rps5 | 2.414356731 |
| ENSRN0P00000026576 | Ribosomal protein S16 | Rps16 | 1.710650338 |
| ENSRN0P00000026696 | Heat shock protein family A member 9 | Hspa9 | 1.471029544 |
| ENSRN0P00000027246 | Ribosomal protein S19 | Rps19 | 2.208197713 |
| ENSRN0P00000027690 | Fibronectin type III domain containing 7 | Fndc7 | 1.23551513 |
| ENSRN0P00000033144 | Ribosomal protein s25 | Rps25 | 2.741821831 |
| ENSRN0P00000033950 | Ubiquitin-like modifier activating enzyme 1 | Uba1 | 1.269082238 |
| ENSRN0P00000034657 | Ubiquitin-like protein fubi and ribosomal protein S30-like | LOC100360647 | 2.548153735 |
| ENSRN0P00000034846 | Heat shock protein 90 beta family member 1 | Hsp90b1 | 1.358989238 |
| ENSRN0P00000038448 | Seryl-tRNA synthetase | Sars | 1.932647244 |
| ENSRN0P00000039630 | Bridging integrator 2 | Bin2 | 1.724795137 |
| ENSRN0P00000042512 | AABR07051533.2 | 1.257161104 | |
| ENSRN0P00000044296 | Actin, beta | Actb | 1.276909267 |
| ENSRN0P00000047328 | AC129049.1 | 1.389931337 | |
| ENSRN0P00000050806 | AABR07065823.2 | 1.240231626 | |
| ENSRN0P00000052173 | Poly(rC) binding protein 2 | Pcbp2 | 1.285290874 |
| ENSRN0P00000056260 | Ribosomal protein S14 | Rps14 | 1.91137709 |
| ENSRN0P00000060949 | Ribosomal protein L34 | Rpl34 | 6.628266907 |
| ENSRN0P00000061700 | AABR07065811.1 | 1.582170833 | |
| ENSRN0P00000061853 | Spectrin, alpha, erythrocytic 1 | Spta1 | 1.245506834 |
| ENSRN0P00000063496 | Annexin A3 | Anxa3 | 1.626219868 |
| ENSRN0P00000064424 | AABR07065778.2 | 1.684985265 | |
| ENSRN0P00000066331 | AABR07065750.2 | 1.720512988 | |
| ENSRN0P00000067217 | Histone cluster 1 H2a family member I like 1 | Hist1h2ail1 | 2.036639138 |
| ENSRN0P00000069086 | AABR07001416.1 | 1.978681684 | |
| ENSRN0P00000070331 | Protein kinase N3 | Pkn3 | 2.193530839 |
| ENSRN0P00000070867 | Neuraminidase 1 | Neu1 | 2.043443611 |
| ENSRN0P00000070868 | Tubulin, alpha 1B | Tuba1b | 1.451033426 |
| ENSRN0P00000071233 | Spectrin, beta, non-erythrocytic 1 | Sptbn1 | 1.49122637 |
| ENSRN0P00000071398 | Glutaminyl-tRNA synthetase | Qars | 1.941751343 |
| ENSRN0P00000072016 | TATA-box binding protein associated factor 15 | Taf15 | 1.442119108 |
| ENSRN0P00000073493 | RAB1A, member RAS oncogene family | Rab1a | 1.726856241 |
| ENSRN0P00000073812 | Kininogen 1 | Kng1 | 2.112182222 |
| ENSRN0P00000074005 | Dyskerin pseudouridine synthase 1 | Dkc1 | 7.605719501 |
| ENSRN0P00000074627 | Multimerin 2 | Mmrn2 | 1.25506891 |
| ENSRN0P00000074688 | Ubiquitin C | Ubc | 1.36418948 |
| ENSRN0P00000075175 | Immunoglobulin heavy constant mu | Ighm | 1.375646382 |
| ENSRN0P00000075269 | Tubulin, beta 5 class I | Tubb5 | 1.260976803 |
| ENSRN0P00000075909 | NFKB activating protein | Nkap | 3.425302711 |
CON: Control.
Figure 6.
GO annotation and KEGG pathway enrichment analysis of differentially expressed proteins between the CON and CUMS groups.
(A) GO annotation of the top 20 differentially expressed proteins. (B) KEGG pathway enrichment analysis. BP: Biological process; CC: cellular components; CON: control; CUMS: chronic unpredictable mild stress; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; MF: molecular function.
DEPs regulated by both CUMS and ICA
Fifty-two DEPs regulated by CUMS and ICA were identified (Figure 7A and Table 1). Among them, 44 DEPs were up-regulated by CUMS and down-regulated by ICA; whereas eight DEPs were down-regulated by CUMS and up-regulated by ICA. GO annotation showed that these 52 DEPs are involved in the formation of cellular components such as cytoplasm, organelles, and cell membranes. They mainly participated in molecular functions such as nucleic acid binding, protein binding, structural composition of ribosomes, and biological processes such as the stress response, biological metabolism, and gene expression (Additional Table 2 (658.7KB, pdf) ). KEGG pathway annotation indicated that these 52 DEPs were mostly involved in the ribosome, phosphatidylinositol 3 kinase (PI3K)/protein kinase B (Akt) signaling, and interleukin-17 (IL-17) signaling pathways (Figure 7B).
Figure 7.
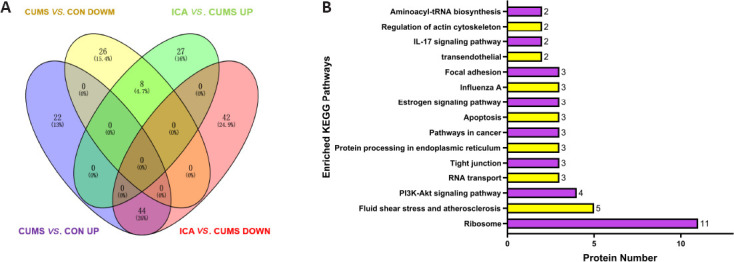
Differentially expressed proteins regulated by both CUMS and ICA.
(A) Venn diagram. (B) KEGG pathway enrichment analysis. CUMS: Chronic unpredictable mild stress; ICA: icariin; KEGG: Kyoto Encyclopedia of Genes and Genomes.
Table 1.
Differentially expressed proteins in the CSF regulated by both CUMS and ICA
| Accession | Protein name | Gene name | CON/CUMS | ICA/CUMS |
|---|---|---|---|---|
| ENSRNOP00000002394 | Chordin | Chrd | 1.330657 | 1.251991 |
| ENSRNOP00000004386 | Myocilin | Myoc | 1.237837 | 1.321273 |
| ENSRN0P00000012286 | UDP-GlcNAc:betaGalbeta-1,3-N-acetylglucosaminyltransferase 2 | B3gnt2 | 1.346536 | 1.214411 |
| ENSRN0P00000016389 | Transforming growth factor, beta induced | Tgfbi | 1.246915 | 1.206508 |
| ENSRN0P00000027860 | HtrA serine peptidase 1 | Htral | 1.246935 | 1.239292 |
| ENSRN0P00000059194 | Angiopoietin-like 1 | Angptll | 1.294498 | 1.220162 |
| ENSRN0P00000070498 | Carboxypeptidase D | Cpd | 1.320809 | 1.340595 |
| ENSRN0P00000071077 | Collagen type II alpha 1 chain | Col2a1 | 1.836686 | 1.680284 |
| ENSRN0P00000001397 | Transmembrane p24 trafficking protein 2 | Tmed2 | 0.473675 | 0.404166 |
| ENSRN0P00000001518 | Ribosomal protein lateral stalk subunit P0 | RplpO | 0.696513 | 0.706795 |
| ENSRN0P00000004278 | Ribosomal protein S4, X-linked | Rps4x | 0.353676 | 0.372436 |
| ENSRN0P00000004867 | Small ubiquitin-like modifier 2 | Sumo2 | 0.746357 | 0.557202 |
| ENSRN0P00000008509 | Eukaryotic translation initiation factor 1A, X-linked | Eiflax | 0.658648 | 0.648145 |
| ENSRN0P00000009249 | Proteasome 26S subunit, non-ATPase 6 | Psmd6 | 0.550629 | 0.486541 |
| ENSRN0P00000009556 | Heat shock protein HSP 90-alpha | HSP90AA1 | 0.730779 | 0.665003 |
| ENSRN0P00000009649 | Proteasome 26S subunit, ATPase 6 | Psmc6 | 0.718329 | 0.621687 |
| ENSRN0P00000010674 | Tyrosyl-transfer RNA synthetase | Yars | 0.556245 | 0.649364 |
| ENSRN0P00000013375 | Eukaryotic translation initiation factor 2 subunit alpha | Eif2sl | 0.465363 | 0.499192 |
| ENSRN0P00000015598 | RAB11a, member RAS oncogene family | Rablla | 0.730942 | 0.70133 |
| ENSRN0P00000017234 | Heparin binding growth factor | Hdgf | 0.771831 | 0.595331 |
| ENSRN0P00000019162 | Ribosomal protein L35 | Rpl35 | 0.61251 | 0.58689 |
| ENSRN0P00000019247 | Ribosomal protein L27a | Rpl27a | 0.673251 | 0.666698 |
| ENSRN0P00000021048 | Myosin light chain 12A | Myl12a | 0.495839 | 0.518126 |
| ENSRN0P00000022184 | Ribosomal protein S12 | Rps12 | 0.255554 | 0.234153 |
| ENSRN0P00000022603 | Calmodulin 1 | Calml | 0.636832 | 0.5501 |
| ENSRN0P00000023935 | Ribosomal protein S3 | Rps3 | 0.519405 | 0.502597 |
| ENSRN0P00000024430 | Vimentin | Vim | 0.727629 | 0.691827 |
| ENSRN0P00000025217 | Ribosomal protein L17 | Rpl17 | 0.482482 | 0.50053 |
| ENSRN0P00000026528 | Ribosomal protein S5 | Rps5 | 0.414189 | 0.469701 |
| ENSRN0P00000026696 | Heat shock protein family A member 9 | Hspa9 | 0.679796 | 0.621869 |
| ENSRN0P00000027246 | Ribosomal protein S19 | Rps19 | 0.452858 | 0.524735 |
| ENSRN0P00000033144 | Ribosomal protein s25 | Rps25 | 0.364721 | 0.310551 |
| ENSRN0P00000033950 | Ubiquitin-like modifier activating enzyme 1 | Uba1 | 0.787971 | 0.715948 |
| ENSRN0P00000034657 | Ubiquitin-like protein fubi and ribosomal protein S30-like | LOC100360647 | 0.392441 | 0.374892 |
| ENSRN0P00000034846 | Heat shock protein 90 beta family member 1 | Hsp90b1 | 0.735841 | 0.802234 |
| ENSRN0P00000038448 | Seryl-transfer RNA synthetase | SerRS | 0.517425 | 0.483297 |
| ENSRN0P00000044296 | Actin, beta | Actb | 0.783141 | 0.768301 |
| ENSRN0P00000056260 | Ribosomal protein S14 | Rps14 | 0.523183 | 0.572311 |
| ENSRN0P00000060949 | Ribosomal protein L34 | Rpl34 | 0.150869 | 0.172269 |
| ENSRN0P0000006442 | - | AABR07065778.2 | 0.593477 | 0.614344 |
| ENSRN0P0000006633 | - | AABR07065750.2 | 0.581222 | 0.541466 |
| ENSRN0P00000067217 | Histone cluster 1 H2a family member I like 1 | Hist1h2ail1 | 0.491005 | 0.388903 |
| ENSRN0P00000070331 | Protein kinase N3 | Pkn3 | 0.455886 | 0.407136 |
| ENSRN0P00000070868 | Tubulin, alpha 1B | Tuba1b | 0.689164 | 0.596259 |
| ENSRN0P00000071233 | Spectrin, beta, non-erythrocytic 1 | Sptbn1 | 0.670589 | 0.628294 |
| ENSRN0P00000072016 | TATA-box binding protein associated factor 15 | Taf15 | 0.693424 | 0.622737 |
| ENSRN0P00000073493 | RAB1A, member RAS oncogene family | Rab1a | 0.579087 | 0.532868 |
| ENSRN0P00000074005 | Dyskerin pseudouridine synthase 1 | Dkc1 | 0.13148 | 0.139895 |
| ENSRN0P00000074688 | Ubiquitin C | Ubc | 0.733036 | 0.750576 |
| ENSRN0P00000075909 | NFKB activating protein | Nkap | 0.291945 | 0.260421 |
CON: Control; CSF: cerebrospinal fluid; CUMS: chronic unpredictable mild stress; ICA: icariin.
Quantification of target proteins by PRM
On the basis of the results from the GO annotation and KEGG enrichment analysis, 10 cell proliferation-related DEPs (Rps3, Rps12, Rps4x, Rps14, Rps19, Hsp90b1, Hsp90aa1, Calm1, Cpd, and HtrA1) were selected for analysis by PRM. The results showed that Rps4x, Rps12, Rps14, Rps19, Hsp90b1, and Hsp90aa1 were up-regulated by CUMS and down-regulated by ICA; whereas HtrA1 was down-regulated by CUMS and up-regulated by ICA (Table 2). The PRM results for seven of these 10 proteins were consistent with the TMT results, thereby confirming the reliability of the TMT results. The TMT results for Rps3, Calm1, and Cpd were not validated by the PRM results.
Table 2.
PRM quantitative analysis of target proteins
| Protein name | Gene name | PRM results | TMT results | ||
|---|---|---|---|---|---|
|
|
|
||||
| CUMS/CON | ICA/CUMS | CUMS/CON | ICA/CUMS | ||
| HtrA serine peptidase 1 | Htra1 | 0.3347 | 1.7893 | 0.8020 | 1.2393 |
| Ribosomal protein S4, X-linked | Rps4x | 3.4306 | 0.8016 | 2.8274 | 0.3724 |
| Heat shock protein HSP 90-alpha | Hsp90aa1 | 3.6841 | 0.3606 | 1.3684 | 0.6650 |
| Ribosomal protein S12 | Rps12 | 6.4356 | 0.4176 | 3.9131 | 0.2342 |
| Ribosomal protein S19 | Rps19 | 11.4161 | 0.1136 | 2.2082 | 0.5247 |
| Heat shock protein 90 beta family member 1 | Hsp90b1 | 3.4992 | 0.4571 | 1.3590 | 0.8022 |
| Ribosomal protein S14 | Rps14 | 15.7618 | 0.0882 | 1.9114 | 0.5723 |
CON: Control; CUMS: chronic unpredictable mild stress; ICA: icariin; PRM: parallel reaction monitoring; TMT: tandem mass tag.
Discussion
Depression is highly prevalent in the general population and is associated with grave consequences, including excessive mortality, disability, secondary morbidity, and high socioeconomic costs. However, the efficacy of current anti-depressants is inadequate, as almost 40% of patients do not recover following an antidepressant trial (Huang et al., 2020). Research is underway to explore the pathogenesis of depression in an attempt to develop more effective and safer anti-depressants. There is increasing interest in the anti-depressive effects of natural compounds, due to their low toxicity and diverse biological properties. In this study we show that ICA exerts significant anti-depressant efficacy in a rat model of depression, alleviating typical depressive symptoms such as anhedonia, decreased locomotor activity, and despair.
The hippocampus is vulnerable to damage from a variety of psychological stressors (Oitzl et al., 1998). Clinically, depressed individuals exhibit cognitive impairments such as memory and learning deficits, implicating hippocampal dysfunction. Dysfunctional hippocampal neurogenesis is one of the main mechanisms underlying depression. Therefore, alleviating hippocampal damage is a current focus in anti-depressant research. In this study, we found that treatment with ICA significantly reduced hippocampal damage, learning and memory impairment, dysfunctional hippocampal neurogenesis, and DG neuronal death in a rat model of depression.
ICA cannot effectively cross the blood-brain barrier or accumulate in the brain (Xu et al., 2017). Therefore, it is unclear how it protects against depression and hippocampal damage. The CSF is the pivotal element that connects the CNS with the periphery and other brain regions, and as such may contain substances that directly regulate hippocampal neurogenesis. We therefore hypothesized that ICA protects the hippocampus from damage and promotes neuronal survival by altering CSF components.
To test this hypothesis in vitro, we simulated chronic stress–induced damage by exposing NSCs to a high concentration of CORT. Exposure to CORT significantly inhibited NSC proliferation and differentiation. CSF from CUMS rats could not prevent CORT-induced injury, while CSF from ICA rats could effectively prevent this damage. These results suggest that ICA alters some CSF components, thereby regulating NSC proliferation and differentiation.
Proteins are the executors of life activities and biological functions. Therefore, changes in CSF proteins may be involved in the effects of ICA on NSC proliferation and differentiation. In this study, we identified 52 DEPs in CSF that were co-regulated by CUMS and ICA. Among these proteins, 44 were up-regulated in response to CUMS exposure and down-regulated after ICA treatment, while eight were down-regulated in response to CUMS exposure and up-regulated after ICA treatment.
GO annotation and KEGG pathway enrichment analysis of these DEPs indicated that three possible pathways are involved in the effects of ICA on depression, dysfunctional neurogenesis, and neuronal death, namely the ribosome, PI3K-Akt signaling, and IL-17 signaling pathways. To confirm the results from the TMT analysis, PRM quantitative analysis was conducted on 10 DEPs that were closely connected with cell proliferation and survival. The PRM results for seven of the 10 DEPs (Rps4x, Rps12, Rps14, Rps19, Hsp90b1, Hsp90aa1 and Htra1) were consistent with the TMT results.
Proteins related to the ribosome pathway
Among the 52 DEPs co-regulated by CUMS and ICA, 11 were enriched in the ribosome pathway, which was the most enriched KEGG pathway. All 11 of these proteins were up-regulated in the CSF by CUMS.
Many studies have confirmed that abnormal ribosomal protein expression or transcription is present in patients with depression and animal models of depression. Most of these studies have observed that ribosomal transcripts or proteins are up-regulated in the blood, liver, hippocampus, and other tissues of depressed individuals (Li et al., 2017; Hori et al., 2018; Guo et al., 2020). In addition, ribosomal protein expression in the hippocampus of rats was significantly down-regulated after 3 weeks of exposure to CUMS (Zhang et al., 2019). According to our previous study (Huang et al., 2020), although rats showed depression-like behaviors after 3 weeks of exposure to CUMS, they were still in the stress compensation stage, and as such did not have hippocampal damage. Different brain regions may respond to mental stress at different times and may react differently towards various stressors. Nevertheless, abnormal expression of ribosomal transcripts and proteins is found in both central and peripheral regions of depressed individuals.
It is worth noting that the PRM analysis showed more significant differences in the expression of ribosomal proteins than did the TMT analysis. This suggests that these ribosomal proteins may play an important role in the development and remission of depression.
Ribosomes are composed of ribosomal RNAs and ribosomal proteins, and are the main sites for intracellular RNA translation, which controls protein synthesis. In response to stress process, ribosomal assembly and protein synthesis increase as the demand for additional protein to cope with stress-induced damage increases (Spriggs et al., 2010; Vadivel Gnanasundram and Fåhraeus, 2018; Wu et al., 2019). In this study, we found that more DEPs were up-regulated than down-regulated in the CSF of CUMS rats.
However, ribosomal assembly requires extremely high rates of coordinated synthesis and assembly of macromolecules across cellular compartments. Defects in ribosomal synthesis may occur during the assembly process, resulting in the rapid accumulation of ribosomal proteins (Warner, 1999; Lempiäinen and Shore, 2009; Tye et al., 2019). In addition, numerous studies have shown that multiple cellular stresses act directly on the nucleus to trigger the overexpression of ribosomal proteins. Therefore, the overexpression of ribosomal proteins may indicate stress-induced ribosomal assembly dysfunction (Zhang and Lu, 2009; Zhou et al., 2012).
Ribosomes control the translation of all proteins, and normal ribosomal synthesis is essential for cell survival, growth, and proliferation. Both over-expression and under-expression of ribosomal proteins can disrupt ribosomal synthesis, lead to dysfunctional ribosomal assembly, and cause cell cycle arrest, senescence, or apoptosis (Turi et al., 2019). In addition, accumulation of ribosomal proteins caused by ribosomal synthesis dysfunction can lead to the collapse of protein folding homeostasis, thereby inhibiting cell growth (Lempiäinen and Shore, 2009; Tye et al., 2019). It may also cause cell cycle arrest or apoptosis through extraribosomal functions of ribosomal proteins (Zhou et al., 2015). For example, Rps14 can activate the p53 pathway to inhibit cell proliferation and induce apoptosis (Zhou et al., 2013). In the CNS, normal ribosomal synthesis also plays an important role in neuronal development. Some forms of synaptic plasticity require rapid, local activation of protein synthesis by the ribosomes (Graber et al., 2013). In addition, disruption of ribosomal gene expression may inhibit neuronal proliferation in the DG (Smagin et al., 2016).
In this study, the increased expression of multiple ribosomal proteins (including Rps4x, Rps12, Rps14, Rps19) that was noted in the CSF of a rat model of depression suggested that dysfunctional ribosomal synthesis may have contributed to the dysfunctional neurogenesis and neuronal loss in the DG that were observed. In contrast, treatment with ICA significantly down-regulated the expression of these proteins in the CSF, thus promoting normal ribosomal synthesis, neuronal repair, and NSC proliferation and differentiation.
In summary, dysfunctional ribosomal synthesis may affect cell proliferation and survival by inducing abnormal expression of ribosomal proteins. ICA may protect against dysfunctional neurogenesis and neuronal loss in a rat model of depression by repairing dysfunctional ribosomal synthesis.
Proteins related to the PI3K-Akt and IL-17 signaling pathways
The PI3K-Akt pathway is one of the classical cell cycle regulation pathways, and is crucial to promoting neuronal survival and neurogenesis (Manning and Toker, 2017). It is also a pathway that is effectively targeted by many anti-depressants (Huang et al., 2014; Pazini et al., 2016). Hsp90b1 and Hsp90aa1 are important proteins within the PI3K-Akt pathway, and abnormal expression these two proteins can inhibit PI3K-Akt pathway activity (Ichikawa et al., 2015; Giulino-Roth et al., 2017). Chronic stress can cause overexpression of Hsp90 family proteins in the brains of animal models of depression (Zhang et al., 2020), and overexpression of Hsp90b1, Hsp90aa1, and HIF1α promotes the expression of NDRG2, which plays a key role in negatively regulating PI3K-Akt signaling (Ichikawa et al., 2015).
Hsp90b1 and Hsp90aa1 are also important members of the IL-17 pathway. Hsp90 family proteins functions as a chaperone to facilitate the folding and assembly of its client proteins. Loss of HSP90 chaperone function results in the degradation of its client protein Act1. As Act1 is required for IL-17 signaling, Hsp90 activity is also required for IL-17 signaling (Kim et al., 2016). Epithelial cells, endothelial cells, and glial cells are all IL-17 targets. IL-17 is activated by binding to the receptors of target cells and ultimately promotes the release of large amounts of inflammatory cytokines from target cells, which is one of many causes of neuronal death in individuals with depression (Nadeem et al., 2017). In addition, some pathological phenomena induced by IL-17 can be reversed by inhibiting the activity of Hsp90 family proteins (Pezzulo et al., 2019).
In this study, we found that the PI3K-Akt and IL-17 signaling pathways were enriched, that Hsp90b1 and Hsp90aa1 were overexpressed in the CSF, and that dysfunctional neurogenesis and hippocampal neuronal loss were present in a rat model of depression. This suggests that Hsp90b1 and Hsp90aa1 overexpression may inhibit the PI3K-Akt pathway and activate the IL-17 pathway in the hippocampus, causing dysfunctional neurogenesis and neuronal loss in these rats. The efficacy of ICA in repairing dysfunctional neurogenesis and neuronal loss may be related to the reduction of Hsp90b1 and Hsp90aa1 levels in the CSF.
In addition, though not enriched in the KEGG pathway analysis, some DEPs co-regulated by CUMS and ICA may be involved in repair of dysfunctional neurogenesis and neural reduction. For example, HtrA serine peptidase 1 (HtrA1) is abundantly expressed in astrocytes. This protein, which was down-regulated in response to CUMS, is related to a variety of neurological diseases. HtrA1 mediates transforming growth factor-β hydrolysis and bone morphogenetic protein inhibition to prevent NSC proliferation and differentiation inhibition (Chen et al., 2018). In this study, ICA treatment reversed the CUMS-induced decreased in Htra1 expression in the CSF, as well as dysfunctional neurogenesis and neuronal loss in the DG, suggesting that the ICA may protect against hippocampal damage by regulating the Htra1 content of the CSF.
In conclusion, a 6-week ICA intervention significantly alleviated dysfunctional hippocampal neurogenesis, neuronal loss in the DG, and memory and learning impairment in a rat model of depression. These effects may be related to ICA-mediated regulation of the levels of Rps14, Hsp90b1, Htra1 and other proteins in the CSF. The main pathways involved include the ribosome, PI3K-Akt, and IL-17 pathways. This suggests that ICA may protect the hippocampus by changing protein expression in the CSF.
This study showed that ICA is a potential treatment for depression. However, there were some limitations to this study. The pathways and targets involved in ICA-mediated protection against hippocampal damage that were identified in this study need to be further verified. In addition, given that metabolites generated by the brain can transfer directly into the CSF, it cannot be ruled out that a small amount of ICA crossing the blood-brain barrier may affect metabolism and lead to changes in CSF protein levels.
Additional files:
Additional Table 1: Differentially expressed proteins of cerebrospinal fluid regulated by chronic unpredictable mild stress (CUMS).
Additional Table 2 (658.7KB, pdf) : Gene Ontology annotation of differentially expressed proteins regulated by chronic unpredictable mild stress and icariin.
Gene Ontology annotation of differentially expressed proteins regulated by chronic unpredictable mild stress and icariin
Acknowledgments:
The study was supported by Research Center for Basic Integrative Medicine, Guangzhou University of Chinese Medicine.
Footnotes
Conflicts of interest: The authors declare that they have no conflict of interest.
Financial support: This study was supported by the National Natural Science Foundation of China, No. 81774102 (to LLW). The funding source had no role in study conception and design, data analysis or interpretation, paper writing or deciding to submit this paper for publication.
Institutional review board statement: The study was approved by the Experimental Animal Ethics Committee of Guangzhou University of Chinese Medicine of China in March 2017.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
Open peer reviewer: Larry Baum, University of Hong Kong, China.Additional files:
Funding: This study was supported by the National Natural Science Foundation of China, No. 81774102 (to LLW).
P-Reviewer: Baum L; C-Editor: Zhao M; S-Editor: Yu J, Li CH; L-Editors: Crow E, Song LP; T-Editor: Jia Y
References
- 1.Al Shweiki MR, Oeckl P, Steinacker P, Hengerer B, Schönfeldt-Lecuona C, Otto M. Major depressive disorder: insight into candidate cerebrospinal fluid protein biomarkers from proteomics studies. Expert Rev Proteomics. 2017;14:499–514. doi: 10.1080/14789450.2017.1336435. [DOI] [PubMed] [Google Scholar]
- 2.Bachmann S. Epidemiology of suicide and the psychiatric perspective. Int J Environ Res Public Health. 2018;15:1425. doi: 10.3390/ijerph15071425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen J, Van Gulden S, McGuire TL, Fleming AC, Oka C, Kessler JA, Peng CY. BMP-responsive protease HtrA1 is differentially expressed in astrocytes and regulates astrocytic development and injury response. J Neurosci. 2018;38:3840–3857. doi: 10.1523/JNEUROSCI.2031-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Wang J, Jia X, Tan X, Hu M. Role of intestinal hydrolase in the absorption of prenylated flavonoids present in Yinyanghuo. Molecules. 2011;16:1336–1348. doi: 10.3390/molecules16021336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danzer SC. Depression, stress, epilepsy and adult neurogenesis. Exp Neurol. 2012;233:22–32. doi: 10.1016/j.expneurol.2011.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ditzen C, Tang N, Jastorff AM, Teplytska L, Yassouridis A, Maccarrone G, Uhr M, Bronisch T, Miller CA, Holsboer F, Turck CW. Cerebrospinal fluid biomarkers for major depression confirm relevance of associated pathophysiology. Neuropsychopharmacology. 2012;37:1013–1025. doi: 10.1038/npp.2011.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duman RS, Aghajanian GK. Synaptic dysfunction in depression: potential therapeutic targets. Science. 2012;338:68–72. doi: 10.1126/science.1222939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franceschelli A, Herchick S, Thelen C, Papadopoulou-Daifoti Z, Pitychoutis PM. Sex differences in the chronic mild stress model of depression. Behav Pharmacol. 2014;25:372–383. doi: 10.1097/FBP.0000000000000062. [DOI] [PubMed] [Google Scholar]
- 9.Giulino-Roth L, van Besien HJ, Dalton T, Totonchy JE, Rodina A, Taldone T, Bolaender A, Erdjument-Bromage H, Sadek J, Chadburn A, Barth MJ, Dela Cruz FS, Rainey A, Kung AL, Chiosis G, Cesarman E. Inhibition of Hsp90 suppresses PI3K/AKT/mTOR signaling and has antitumor activity in burkitt lymphoma. Mol Cancer Ther. 2017;16:1779–1790. doi: 10.1158/1535-7163.MCT-16-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graber TE, Hébert-Seropian S, Khoutorsky A, David A, Yewdell JW, Lacaille JC, Sossin WS. Reactivation of stalled polyribosomes in synaptic plasticity. Proc Natl Acad Sci U S A. 2013;110:16205–16210. doi: 10.1073/pnas.1307747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo J, Zhang F, Gao J, Guan X, Liu B, Wang X, Qin Z, Tang K, Liu S. Proteomics-based screening of the target proteins associated with antidepressant-like effect and mechanism of Saikosaponin A. J Cell Mol Med. 2020;24:174–188. doi: 10.1111/jcmm.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hori H, Nakamura S, Yoshida F, Teraishi T, Sasayama D, Ota M, Hattori K, Kim Y, Higuchi T, Kunugi H. Integrated profiling of phenotype and blood transcriptome for stress vulnerability and depression. J Psychiatr Res. 2018;104:202–210. doi: 10.1016/j.jpsychires.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 13.Huang X, Mao YS, Li C, Wang H, Ji JL. Venlafaxine inhibits apoptosis of hippocampal neurons by up-regulating brain-derived neurotrophic factor in a rat depression model. Pharmazie. 2014;69:909–916. [PubMed] [Google Scholar]
- 14.Huang YL, Zeng NX, Chen J, Niu J, Luo WL, Liu P, Yan C, Wu LL. Dynamic changes of behaviors, dentate gyrus neurogenesis and hippocampal miR-124 expression in rats with depression induced by chronic unpredictable mild stress. Neural Regen Res. 2020;15:1150–1159. doi: 10.4103/1673-5374.270414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichikawa T, Nakahata S, Tamura T, Manachai N, Morishita K. The loss of NDRG2 expression improves depressive behavior through increased phosphorylation of GSK3β. Cell Signal. 2015;27:2087–2098. doi: 10.1016/j.cellsig.2015.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Kim BK, Park M, Kim JY, Lee KH, Woo SY. Heat shock protein 90 is involved in IL-17-mediated skin inflammation following thermal stimulation. Int J Mol Med. 2016;38:650–658. doi: 10.3892/ijmm.2016.2627. [DOI] [PubMed] [Google Scholar]
- 17.Lempiäinen H, Shore D. Growth control and ribosome biogenesis. Curr Opin Cell Biol. 2009;21:855–863. doi: 10.1016/j.ceb.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Lepko T, Pusch M, Müller T, Schulte D, Ehses J, Kiebler M, Hasler J, Huttner HB, Vandenbroucke RE, Vandendriessche C, Modic M, Martin-Villalba A, Zhao S, E LL-B, Schneider A, Fischer A, Breunig CT, Stricker SH, Götz M, Ninkovic J. Choroid plexus-derived miR-204 regulates the number of quiescent neural stem cells in the adult brain. EMBO J. 2019;38:e100481. doi: 10.15252/embj.2018100481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li C, Guo Z, Zhao R, Sun W, Xie M. Proteomic analysis of liver proteins in a rat model of chronic restraint stress-induced depression. Biomed Res Int. 2017;2017:7508316. doi: 10.1155/2017/7508316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, Xu C, Wu X, Liu F, Du Y, Sun J, Tao J, Dong J. Icariin exerts an antidepressant effect in an unpredictable chronic mild stress model of depression in rats and is associated with the regulation of hippocampal neuroinflammation. Neuroscience. 2015;294:193–205. doi: 10.1016/j.neuroscience.2015.02.053. [DOI] [PubMed] [Google Scholar]
- 21.Liu L, Zhao Z, Lu L, Liu J, Sun J, Dong J. Icariin and icaritin ameliorated hippocampus neuroinflammation via mediating HMGB1 expression in social defeat model in mice. Int Immunopharmacol. 2019;75:105799. doi: 10.1016/j.intimp.2019.105799. [DOI] [PubMed] [Google Scholar]
- 22.Maller JJ, Daskalakis ZJ, Fitzgerald PB. Hippocampal volumetrics in depression: the importance of the posterior tail. Hippocampus. 2007;17:1023–1027. doi: 10.1002/hipo.20339. [DOI] [PubMed] [Google Scholar]
- 23.Manning BD, Toker A. AKT/PKB signaling: navigating the network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nadeem A, Ahmad SF, Al-Harbi NO, Fardan AS, El-Sherbeeny AM, Ibrahim KE, Attia SM. IL-17A causes depression-like symptoms via NFκB and p38MAPK signaling pathways in mice: Implications for psoriasis associated depression. Cytokine. 2017;97:14–24. doi: 10.1016/j.cyto.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 25.O’Leary OF, Cryan JF. A ventral view on antidepressant action: roles for adult hippocampal neurogenesis along the dorsoventral axis. Trends Pharmacol Sci. 2014;35:675–687. doi: 10.1016/j.tips.2014.09.011. [DOI] [PubMed] [Google Scholar]
- 26.Ogawa S, Tsuchimine S, Kunugi H. Cerebrospinal fluid monoamine metabolite concentrations in depressive disorder: A meta-analysis of historic evidence. J Psychiatr Res. 2018;105:137–146. doi: 10.1016/j.jpsychires.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 27.Oitzl MS, Fluttert M, de Kloet ER. Acute blockade of hippocampal glucocorticoid receptors facilitates spatial learning in rats. Brain Res. 1998;797:159–162. doi: 10.1016/s0006-8993(98)00387-4. [DOI] [PubMed] [Google Scholar]
- 28.Park SC. Neurogenesis and antidepressant action. Cell Tissue Res. 2019;377:95–106. doi: 10.1007/s00441-019-03043-5. [DOI] [PubMed] [Google Scholar]
- 29.Pazini FL, Cunha MP, Rosa JM, Colla AR, Lieberknecht V, Oliveira Á, Rodrigues AL. Creatine, similar to ketamine, counteracts depressive-like behavior induced by corticosterone via PI3K/Akt/mTOR pathway. Mol Neurobiol. 2016;53:6818–6834. doi: 10.1007/s12035-015-9580-9. [DOI] [PubMed] [Google Scholar]
- 30.Pezzulo AA, Tudas RA, Stewart CG, Buonfiglio LGV, Lindsay BD, Taft PJ, Gansemer ND, Zabner J. HSP90 inhibitor geldanamycin reverts IL-13- and IL-17-induced airway goblet cell metaplasia. J Clin Invest. 2019;129:744–758. doi: 10.1172/JCI123524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Planques A, Oliveira Moreira V, Dubreuil C, Prochiantz A, Di Nardo AA. OTX2 signals from the choroid plexus to regulate adult neurogenesis. eNeuro. 2019;6:ENEURO0262–02182019. doi: 10.1523/ENEURO.0262-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qin Y, Jiang X, Li W, Li J, Tian T, Zang G, Fang L, Zhou C, Xu B, Gong X, Huang C, Yang X, Bai M, Fan L, Xie P. Chronic mild stress leads to aberrant glucose energy metabolism in depressed Macaca fascicularis models. Psychoneuroendocrinology. 2019;107:59–69. doi: 10.1016/j.psyneuen.2019.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Skipor J, Thiery JC. The choroid plexus--cerebrospinal fluid system: undervaluated pathway of neuroendocrine signaling into the brain. Acta Neurobiol Exp (Wars) 2008;68:414–428. doi: 10.55782/ane-2008-1708. [DOI] [PubMed] [Google Scholar]
- 34.Smagin DA, Kovalenko IL, Galyamina AG, Bragin AO, Orlov YL, Kudryavtseva NN. Dysfunction in ribosomal gene expression in the hypothalamus and hippocampus following chronic social defeat stress in male mice as revealed by RNA-Seq. Neural Plast. 2016;2016:3289187. doi: 10.1155/2016/3289187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- 36.Turi Z, Lacey M, Mistrik M, Moudry P. Impaired ribosome biogenesis: mechanisms and relevance to cancer and aging. Aging (Albany NY) 2019;11:2512–2540. doi: 10.18632/aging.101922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tye BW, Commins N, Ryazanova LV, Wühr M, Springer M, Pincus D, Churchman LS. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. Elife. 2019;8:e43002. doi: 10.7554/eLife.43002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vadivel Gnanasundram S, Fåhraeus R. Translation stress regulates ribosome synthesis and cell proliferation. Int J Mol Sci. 2018;19:3757. doi: 10.3390/ijms19123757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 40.Wei K, Xu Y, Zhao Z, Wu X, Du Y, Sun J, Yi T, Dong J, Liu B. Icariin alters the expression of glucocorticoid receptor, FKBP5 and SGK1 in rat brains following exposure to chronic mild stress. Int J Mol Med. 2016;38:337–344. doi: 10.3892/ijmm.2016.2591. [DOI] [PubMed] [Google Scholar]
- 41.Wu CC, Zinshteyn B, Wehner KA, Green R. High-resolution ribosome profiling defines discrete ribosome elongation states and translational regulation during cellular stress. Mol Cell. 2019;73:959–970.e5. doi: 10.1016/j.molcel.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu L, Ran C, Liu S, Liao L, Chen Y, Guo H, Wu W, Yan C. Jiaweisinisan facilitates neurogenesis in the hippocampus after stress damage. Neural Regen Res. 2013;8:1091–1102. doi: 10.3969/j.issn.1673-5374.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu LL, Liu Y, Yan C, Pan Y, Su JF, Wu WK. Antidepressant-like effects of fractions prepared from danzhi-xiaoyao-san decoction in rats with chronic unpredictable mild stress: effects on hypothalamic-pituitary-adrenal axis, arginine vasopressin, and neurotransmitters. Evid Based Complement Alternat Med. 2016;2016:6784689. doi: 10.1155/2016/6784689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu S, Yu J, Zhan J, Yang L, Guo L, Xu Y. Pharmacokinetics, tissue distribution, and metabolism study of icariin in rat. Biomed Res Int. 2017;2017:4684962. doi: 10.1155/2017/4684962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang YJ, Li YJ, Li SS, Liu XM, Wang Q. Comparison of experimental maze tests used to assess the learning and memory abilities in rats and mice. Zhongguo Bijiao Yixue Zazhi. 2018;28:129–134. [Google Scholar]
- 46.Zhang J, Zhang Z, Zhang J, Zhong Z, Yao Z, Qu S, Huang Y. iTRAQ-based protein profiling in CUMS rats provides insights into hippocampal ribosome lesion and ras protein changes underlying synaptic plasticity in depression. Neural Plast. 2019;2019:7492306. doi: 10.1155/2019/7492306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y, Lu H. Signaling to p53: ribosomal proteins find their way. Cancer Cell. 2009;16:369–377. doi: 10.1016/j.ccr.2009.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang Y, Du L, Bai Y, Han B, He C, Gong L, Huang R, Shen L, Chao J, Liu P, Zhang H, Zhang H, Gu L, Li J, Hu G, Xie C, Zhang Z, Yao H. CircDYM ameliorates depressive-like behavior by targeting miR-9 to regulate microglial activation via HSP90 ubiquitination. Mol Psychiatry. 2020;25:1175–1190. doi: 10.1038/s41380-018-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou X, Liao JM, Liao WJ, Lu H. Scission of the p53-MDM2 Loop by Ribosomal Proteins. Genes Cancer. 2012;3:298–310. doi: 10.1177/1947601912455200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou X, Hao Q, Liao J, Zhang Q, Lu H. Ribosomal protein S14 unties the MDM2-p53 loop upon ribosomal stress. Oncogene. 2013;32:388–396. doi: 10.1038/onc.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou X, Liao WJ, Liao JM, Liao P, Lu H. Ribosomal proteins: functions beyond the ribosome. J Mol Cell Biol. 2015;7:92–104. doi: 10.1093/jmcb/mjv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu YX, Ye DQ, Li XL. An improved method of collecting cerebrospinal fluid in SD rats. Zhongguo Bijiao Yixue Zazhi. 2018;28:113–115. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene Ontology annotation of differentially expressed proteins regulated by chronic unpredictable mild stress and icariin