Extended Data Fig. 7 |. PRC1 does not restrict BAF binding despite extensive overlap.
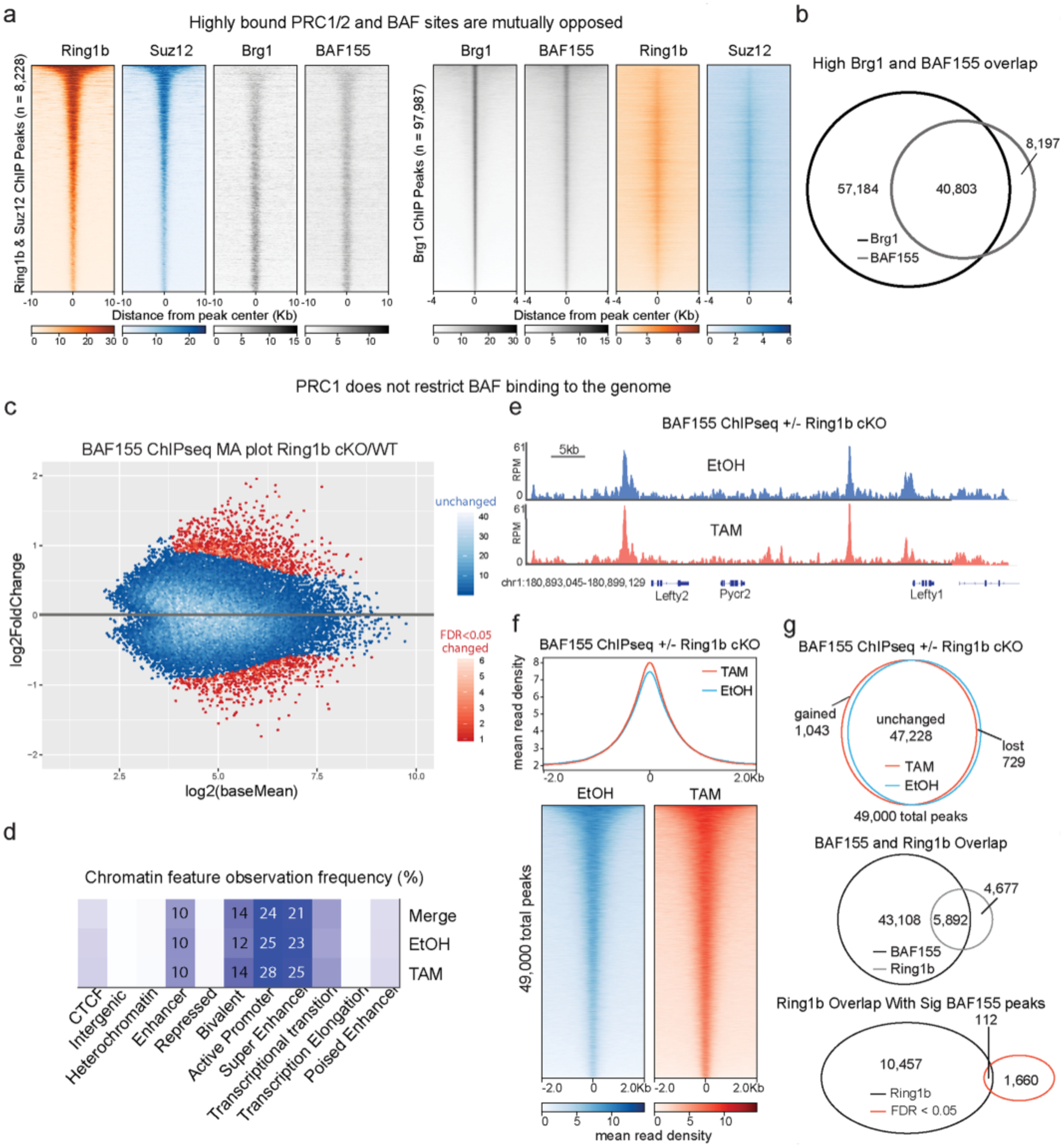
a, Heatmaps depicting Ring1b, Suz12, BAF155, and Brg1 ChIPseq signal from79 at peaks (8,228) that overlap Ring1b and Suz12, sorted by decreasing Ring1b signal (left) or Brg1 peaks (97,987) sorted by decreasing Brg1 signal (right). b, High overlap between Brg1 and BAF155 despite different number of peaks called due to antibody quality and read depth. c, BAF155 ChIP-seq in Ring1bfl/fl × Ring1a−/− × ActinCreER ESCs treated with Tamoxifen or EtOH for 72 h. MA plot (TAM/EtOH) highlights large number of unchanged peaks (>95% in blue) (n = 2). d, Analysis of the chromatin features that characterize BAF155 binding sites reveal that Ring1a/b double KO does not alter BAF complex binding, as the complex remains mainly bound to enhancers and bivalent promoters in both control and mutant ESCs. e, Representative genome tracks showing similar BAF155 levels across the Lefty1/2 locus in Tamoxifen and EtOH treated cells. f, Metagene plot and heatmaps showing that BAF155 ChIPseq signal is minimally affected by Ring1b deletion (g) Characterization of BAF155 peaks in EtOH and Tamoxifen conditions. Of the 49,000 detected BAF155 peaks in this dataset, less than 5% were significantly changed upon Ring1a/b double KO in ESCs and the small number of gained and decreased sites was similar. Despite 56% of Ring1b peaks overlapping with a BAF peak, only 1% of differentially bound peaks are within a PRC1 domain (bottom).
