Extended Data Fig. 8 |. Characterization of EED/Ring1b degron and extended analysis related to Fig. 5.
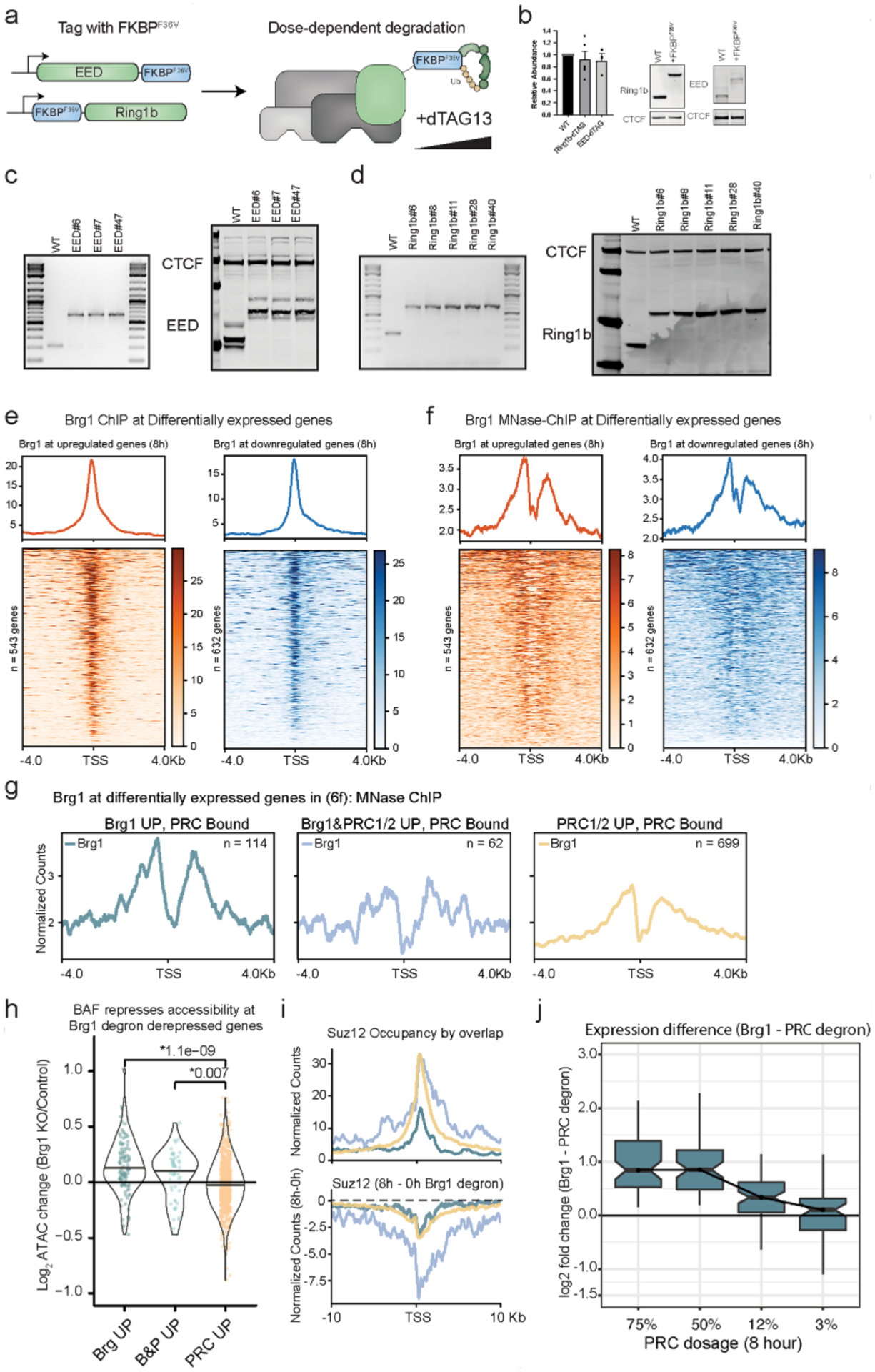
a, Schematic depicting EED and Ring1b dTAG targeted protein degradation strategy in mESCs, where EED is tagged at the C-terminus and Ring1b is tagged at the N-terminus with FKBPF36V to enable targeted degradation with dTAG13 PROTAC. b, Representative western blot and quantitation of EED and Ring1b abundance in parent line compared to lines tagged with FKBPF36V. Error bars depict mean ± standard error of the mean from three tagged EED lines and four tagged Ring1b lines. c, Genotyping PCR and western from whole cell extract showing 14.6kDa shift in migration from C-terminus (G4S)3-FKBPF36V-G4S-V5 tag on three different EED clones used for experiments. d, Genotyping PCR and western from whole cell extract with 14.2kDa shift in migration from N-terminus FKBPF36V-G4S-HA-(G4S)3. Experiment was repeated at least 2 times with similar results (e) Brg1 ChIPseq showing localization to the TSS of differentially expressed genes, ChIP from79. f, Brg1 MNase ChIPseq showing localization to the TSS of differentially expressed genes, ChIP from80. g, Brg1 MNase ChIPseq showing higher occupancy at PRC bound genes repressed by Brg1 (independent of polycomb). Compare to Fig. 5f. h, Brg1 represses accessibility at PRC bound genes derepressed by Brg1 degron and Brg1/PRC degron. Accessibility change (Brg1 KO/control) by overlap (Fig. 6f) for peaks ± 2kb from gene promoter, ATAC data from44 with two-sided t-test p-value above. (i,top) Higher and broader polycomb domains over genes derepressed by both Brg1 and PRC degron than genes derepressed by Brg1 degron alone, with Suz12 occupancy shown here, (i, bottom) Broad domains over genes derepressed by both Brg1 and PRC degron show substantially more redistribution in the Brg1 degron. Compare to Ring1b signal in Fig. 6. (j) Expression difference (Brg1 - PRC Log2 Fold Change) for genes derepressed by Brg1 and PRC degron, over dosage titration (Most dosage sensitive genes are shown in Fig. 6g), from n = 4 biological replicates. Boxplot bounds depict quartile 1, median, and quartile 3 with whiskers at 1.5× interquartile range.
