Fig. 5 |. BAF promotes repression directly and by genome-wide Polycomb redistribution.
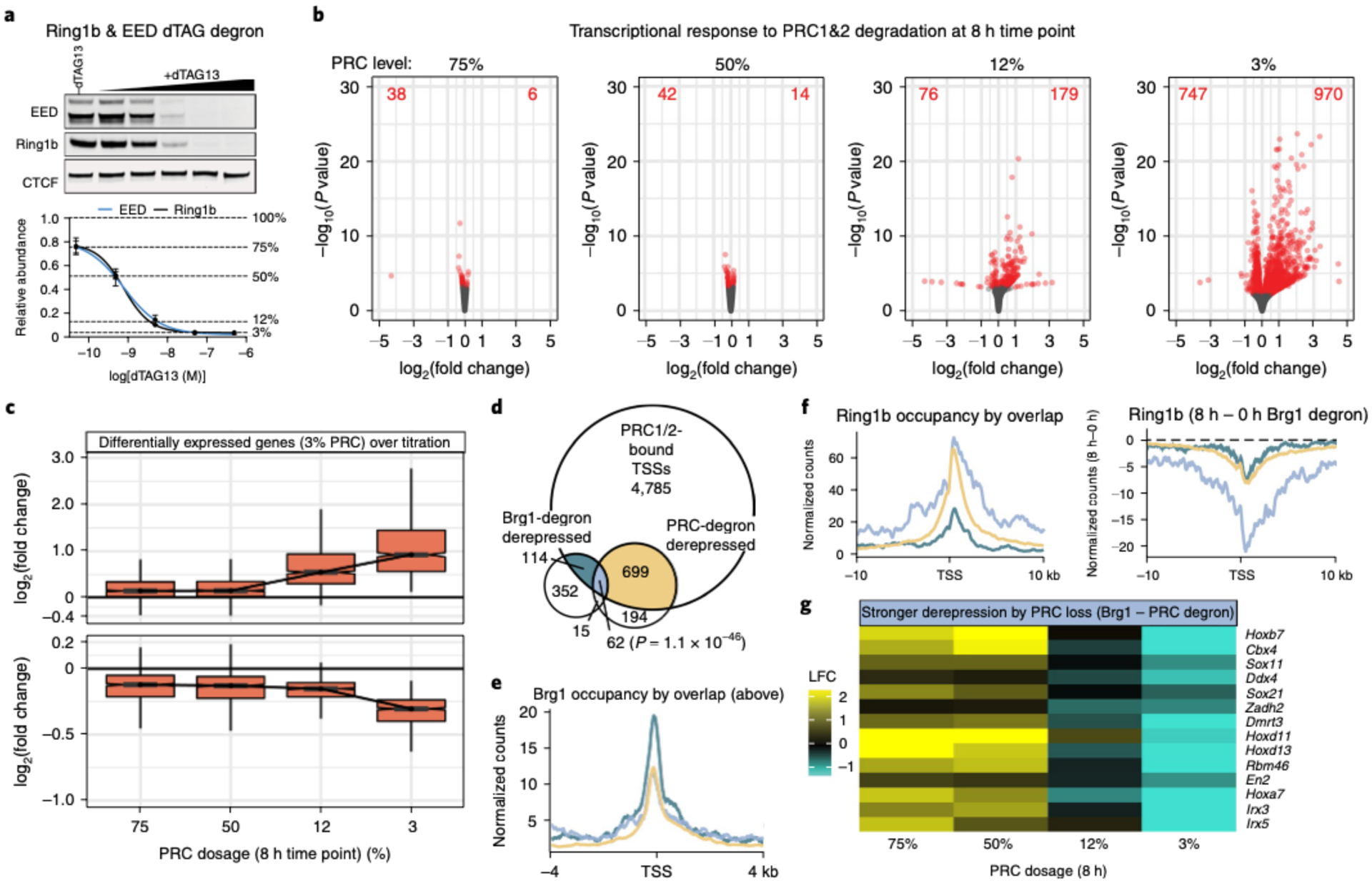
a. Representative western blot (top) and quantification (bottom) of Ring1b (PRC1) and EED (PRC2) degron lines across 10-fold dilution treatment with dTAG13 proteolysis-targeting chimera (PROTAC) for 8 h. Data are presented as mean ± s.e.m. from four cell lines. b, Transcriptional response to degrading PRC1/2 to 75, 50, 12 and 3% of WT levels for 8 h. The volcano plots depict gene expression changes across four PRC doses, with differentially expressed genes colored red (FDR-corrected P < 0.05) from n = 4 cell lines. c, Fold change in expression level (log2) for genes that are differentially expressed at 3% PRC dosage (FDR-corrected P < 0.05) across all doses (increased n = 970 (top), decreased n = 747 (bottom)). The boxplot bounds depict quartile 1, median and quartile 3, with whiskers at 1.5× interquartile range. d, Plot showing significant overlap between genes that are bound by PRC1/2 (Ring1b and Suz12 ± 2kb from TSS), Brg1-degron derepressed (8 h) and PRC1/2-degron derepressed (3% dosage, 8 h). The significance of the overlap of PRC-bound, Brg1 and PRC upregulated genes was calculated by Fisher’s exact test (P = 1.1 × 10−46). e, Brg1 occupancy is substantially higher over PRC1/2 bound genes solely derepressed in the Brg1 degron compared to genes derepressed by PRC or PRC and Brg1 degradation. Colors are as in d. f, Left: Polycomb domains are higher and broader over genes derepressed by both Brg1 and PRC degron than genes derepressed by Brg1 degron alone. Right: broad domains over genes derepressed by both Brg1 and PRC degron show substantially more redistribution in the Brg1 degron. Colors are as in d. g, Heatmap depicting the log2 fold change (LFC) in expression between the Brg1 degron and each PRC degron dose for select genes more strongly derepressed by PRC than Brg1 degradation. All experiments were conducted in serum/LIF.
