Extended Data Fig. 1|. Extended characterization of Brg1-AID degron, related to Fig. 1.
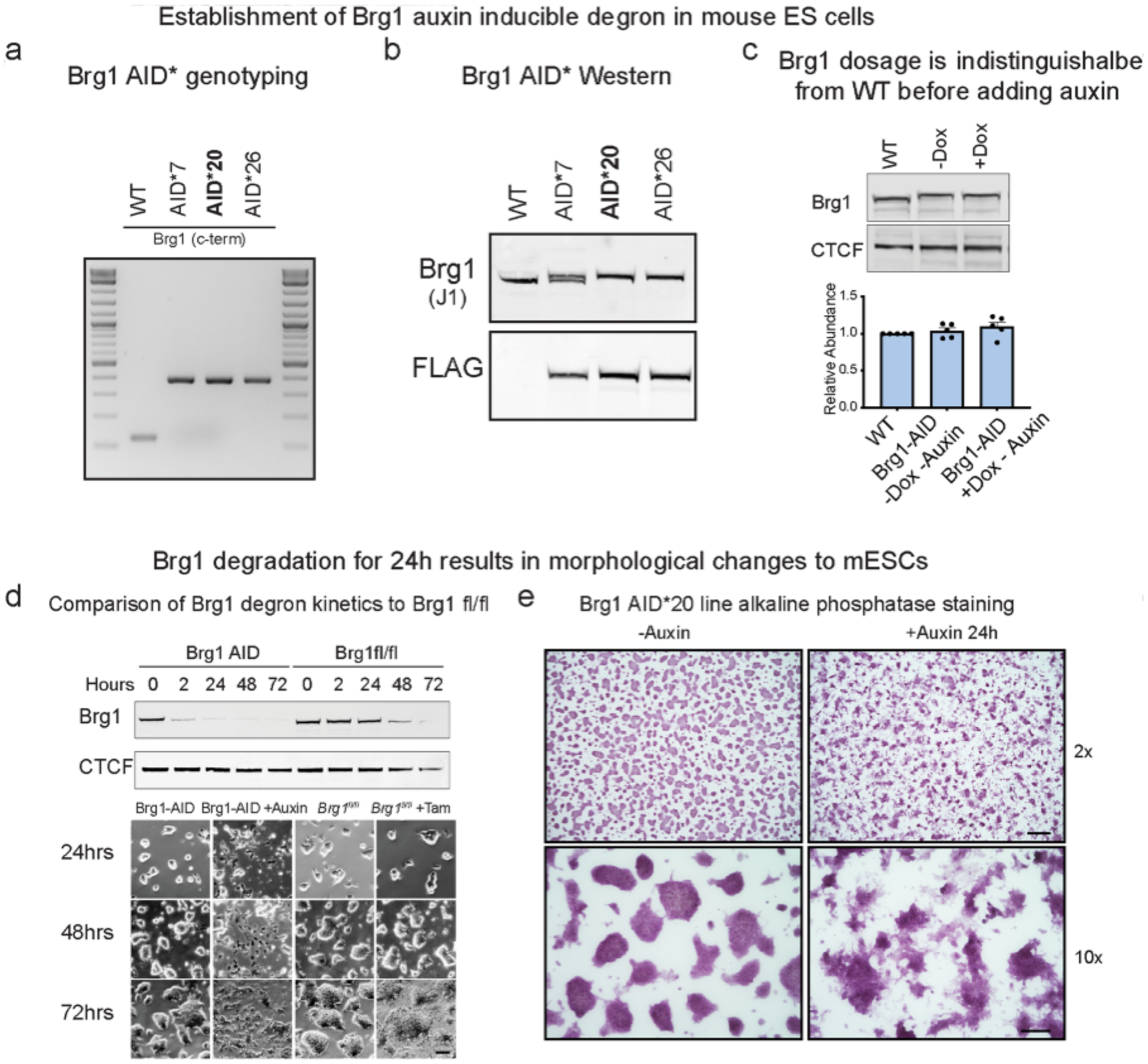
(a, Genotyping PCR for three homozygous knock in clones (AID*20 was used for most experiments in the manuscript). b, Western blot of whole-cell extracts from knock-in lines showing 8.8kDa shift in migration from c-terminal tag (G4S)3-AID*-G4S-3xFLAG. Line 20 and 26 have homozygous insertions in frame that were also confirmed by Sanger sequencing. c, Representative western blot and quantitation of Brg1 abundance in parent line compared to Brg1-AID* ± osTIR1 induction (-auxin). Error bars represent mean ± standard deviation from five biological replicates. d, Comparison of Brg1 depletion kinetics between AID and Cre-lox genetic deletion by western blot and changes to colony morphology that occur at the 24-hour time-point. Cells were only grown on the same dish for 72 h to illustrate timing for Brg1fl/fl colony morphology changes. Experiment was repeated at least 3 times with similar results. Scale bar = 50 μm. e, Alkaline phosphatase staining of Brg1 AID* 20 line on gelatin coated dishes, showing that changes to colony morphology occur ~24 h. Top scale bar = 200 μm, bottom scale bar = 50μm. Experiment was repeated at least 3 times with similar results.
