Fig. 3.
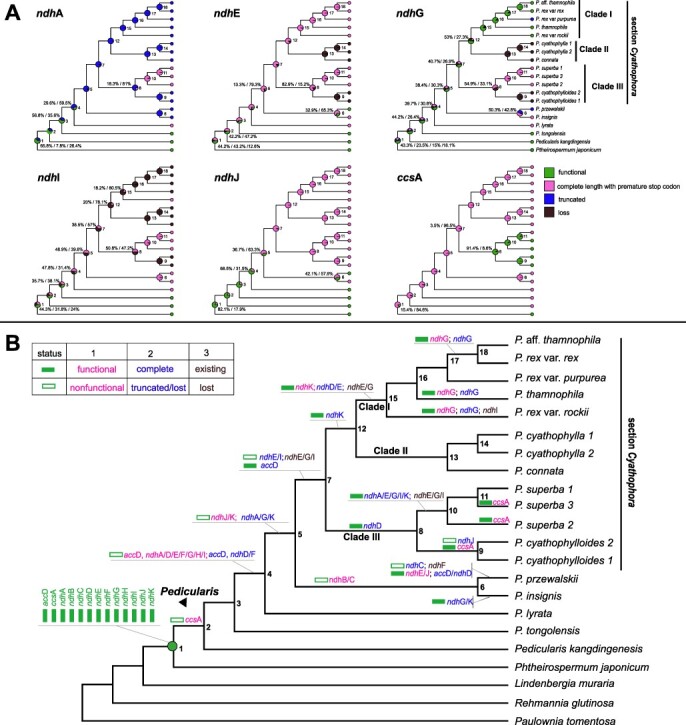
Ancestral state reconstruction of accD, ccsA and NDH genes in Pedicularis based on the ‘ER’ likelihood model using ‘phytools’ package in R program (A) and parsimony method using Dollop program in the PHYLIP packages (B). Important nodes are annotated by numbers. Genes were classified as four states, i.e. functional (0), full sequence with premature stop codon (1), truncated gene (2) and complete loss (3). In the ‘ER’ model analyses, we inferred ancestral states using four states. In the Dollop analyses, we classified three types: (1) functional (state 0) or non-functional (state 1/2/3), (2) complete (state 0/1) or truncated/lost (state 2/3) and existing (state 0/1/2) or lost (state 3), then we inferred ancestral states using two states.
