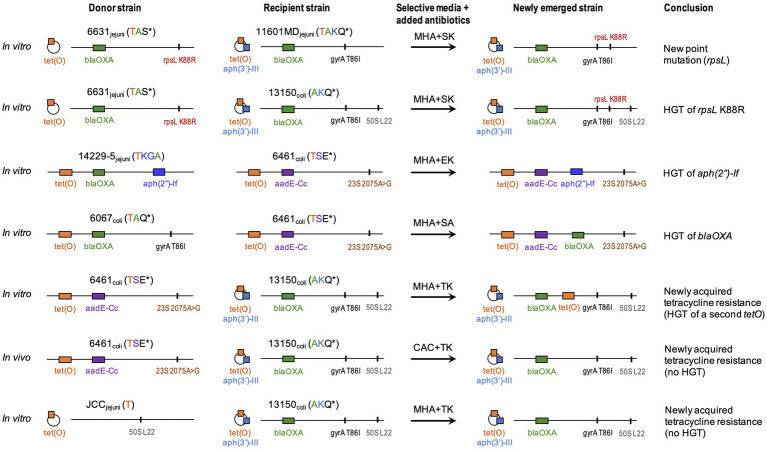
Figure 1.
Summary of in vitro and in vivo experiments. Strains names are followed by Campylobacter species and phenotypic resistance. Acronyms include A, ampicillin/carbenicillin; E, erythromycin; G, gentamicin; K, kanamycin; Q, nalidixic acid; S, streptomycin; and T, tetracycline. *indicates resistances due to a point mutation. Genotypic resistance can be carried on a chromosome (horizontal line) or a plasmid (circle). Resistance genes (colored blocks) are as: tet(O) (T resistance); aph(3′)-IIIa (K resistance); blaOXA (A resistance); aadE-Cc (S resistance); and aph(2′)-If (KG resistance). Point mutations (vertical lines) are as: gyrA T86I (nalidixic acid/ciprofloxacin: Q resistance); 23S A2075G (E resistance); rpsL K88R (S resistance); and L22 A103V (low macrolide resistance). Whole genome sequence data from newly emerged strain and parental strains were compared to identify donor vs recipient. CAC, CHROMagar™ Campylobacter; MHA, Mueller-Hinton agar; and HGT, horizontal gene transfer. Strain 13150 (phenotypically susceptible to tetracycline) carries a plasmid-borne non-functional tet(O) represented as a striped orange block.