Figure 5.
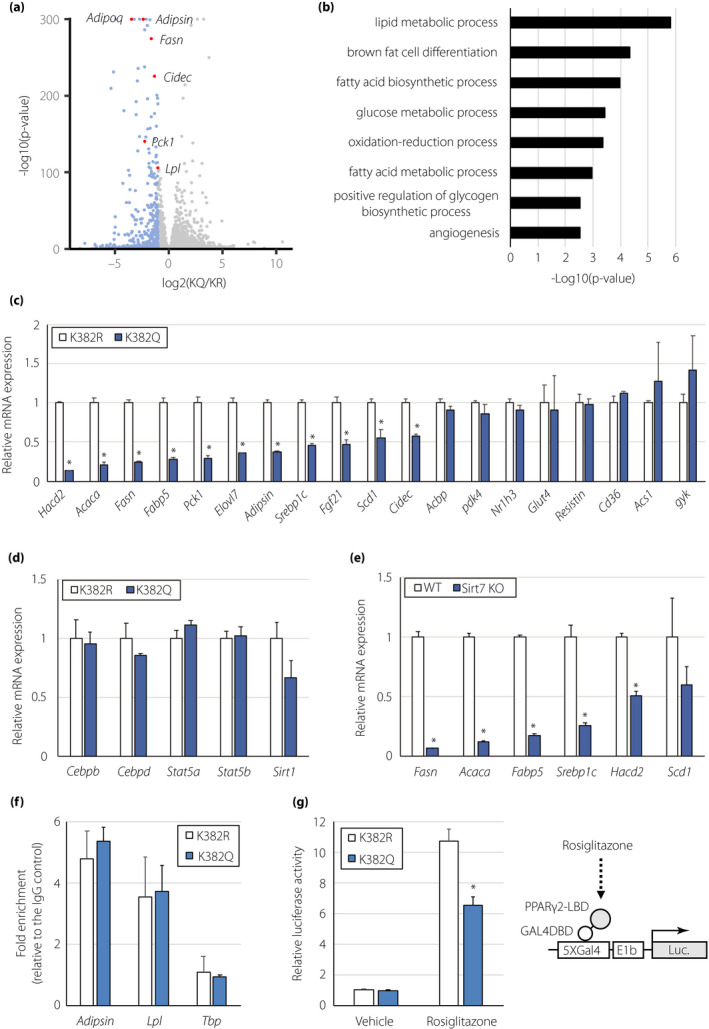
PPARγ2 acetylation at K382 regulates the expression of lipogenesis‐related genes. (a) Volcano plot derived from RNA‐seq analysis of PPARγ2K382R‐ and PPARγ2K382Q‐expressing adipocytes. Transcripts downregulated (fold change > 2, P < 0.05) in PPARγ2K382Q‐expressing adipocytes are in blue. (b) Gene ontology analysis of the downregulated genes in PPARγ2K382Q‐expressing cells. (c, d) Expression of genes involved in lipid metabolism (c) and adipocyte differentiation (d) in PPARγ2K382R‐ and PPARγ2K382Q‐expressing adipocytes (n = 3). (e) Expression of lipogenic genes in epiWAT of WT and Sirt7 KO mice (n = 4). (f) ChIP for the recruitment of PPARγ to the indicated genes in PPARγ2K382R‐ and PPARγ2K382Q‐expressing adipocytes (n = 3). Quantification of enrichment is represented as fold‐enrichment relative to IgG. (g) Effect of K382 acetylation on the ligand‐dependent activity of PPARγ2 in HEK293T cells. The cells were transfected with the GAL4DBD‐PPARγ2 LBDK382R or GAL4DBD‐PPARγ2 LBDK382Q expression plasmid, as well as the 5×GAL4‐luciferase reporter plasmid, followed by treatment with or without rosiglitazone. Luciferase activity was determined after 18 h (n = 4). Data are shown as the mean ± the standard error of the mean. *P < 0.05
