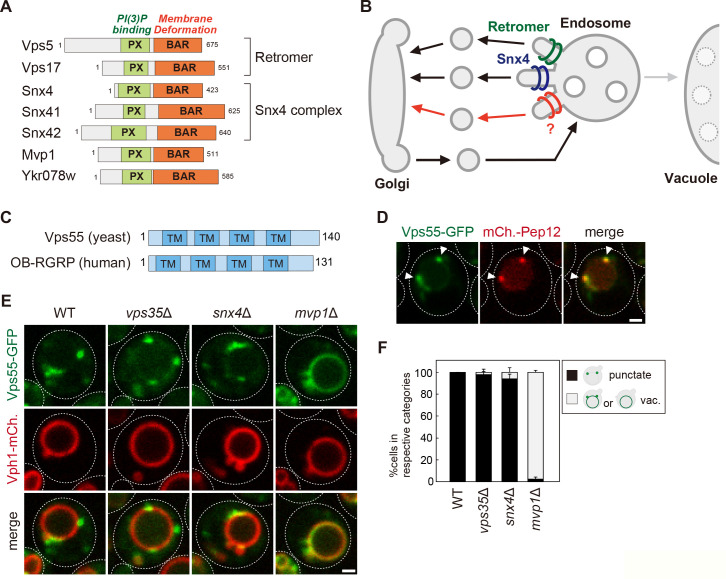
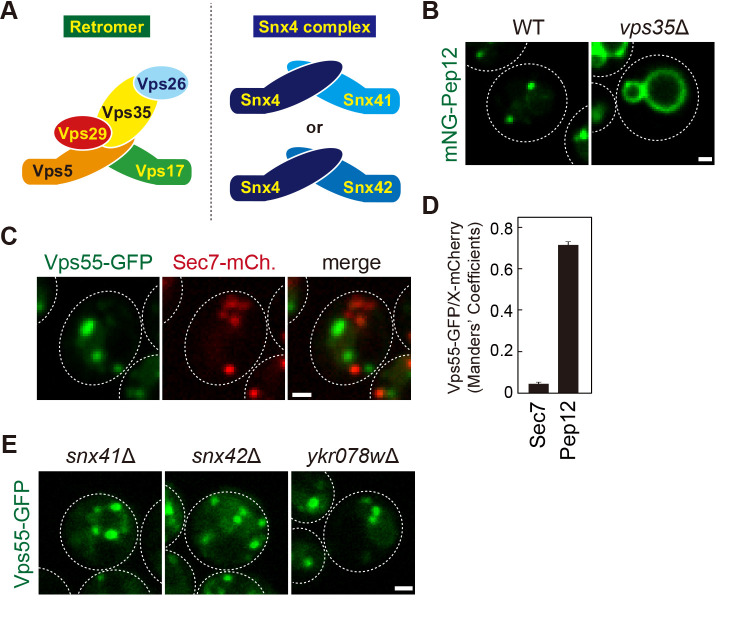
Figure 1. The endosomal localization of Vps55 requires Mvp1.
(A) Schematic of SNX-BAR proteins in yeast. (B) Model of endosomal recycling pathways in yeast. (C) Schematic of Vps55 and OB-RGRP. (D) Vps55-GFP localization. The mCherry-Pep12 serves as an endosomal marker. (E) Vps55-GFP localization in wild-type (WT), vps35Δ (retromer mutant), snx4Δ (Snx4 complex mutant), and mvp1Δ. (F) Quantification of Vps55-GFP localization from three independent experiments. N = >30 cells. Scale bar: 1 µm.