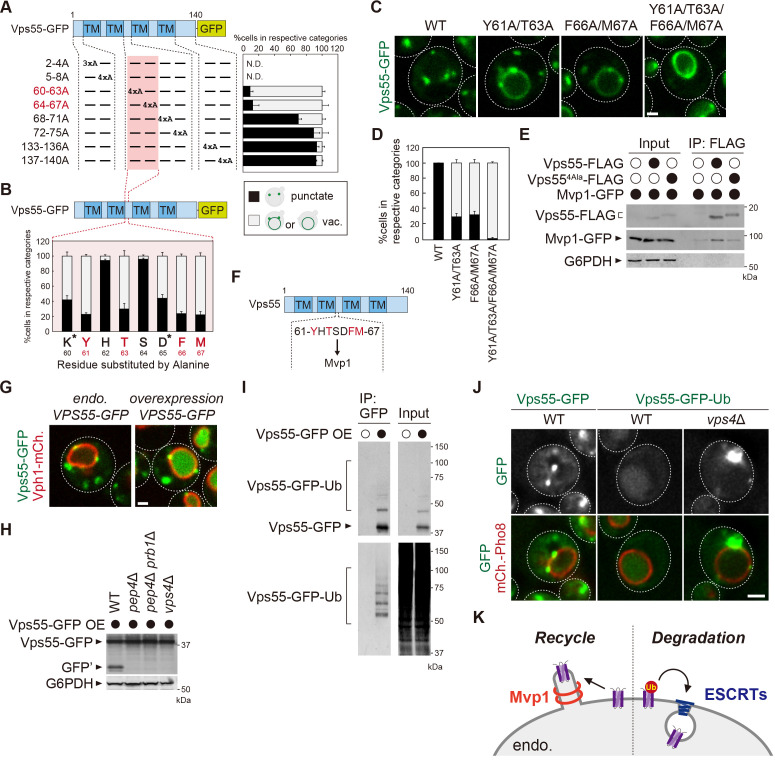
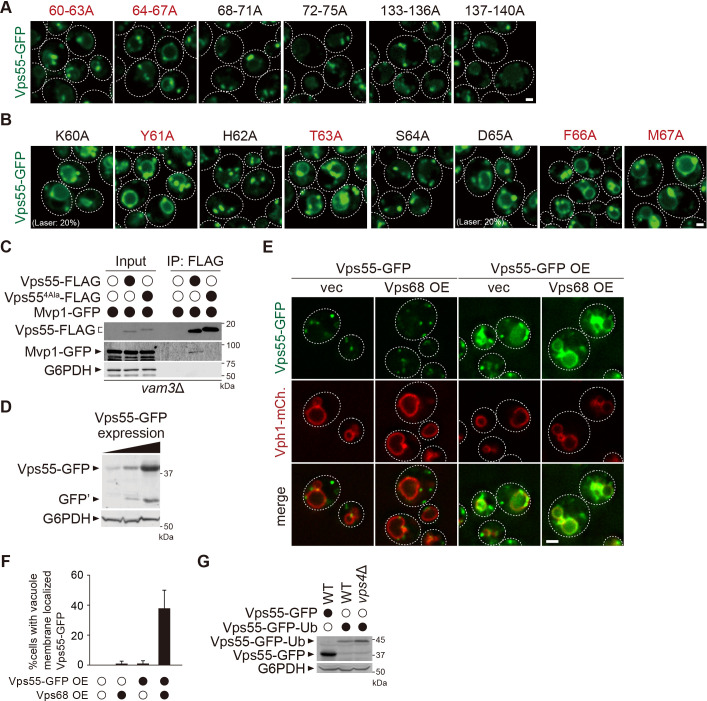
Figure 3. Mvp1 recognizes Vps55 through a specific sorting motif.
(A, B, and D) Schematic of Vps55 mutational analysis and quantitation of Vps55-GFP mutant localization, from Figure 3—figure supplement 1A (A), Figure 3—figure supplement 1B (B), and Figure 3—figure supplement 1C (D). (C) The localization of Vps55-GFP mutants. (E) The Mvp1-Vps55 interaction in Vps55-FLAG mutants. Vps55-FLAG mutants were immunoprecipitated (IP) from cells expressing Mvp1-GFP, and the IP products were analyzed by immunoblotting using antibodies against FLAG, green fluorescent protein (GFP), and glucose-6-phosphate dehydrogenase (G6PDH). (F) Schematic of Vps55 and the residues facilitating its interaction with Mvp1. (G) The localization of overexpressed Vps55-GFP. (H) Vps55-GFP sorting in vacuolar hydrolases (pep4Δ and pep4Δ prb1Δ) and ESCRT (vps4Δ) mutants. Cell lysates expressing Vps55-GFP were analyzed by immunoblotting using antibodies against GFP and G6PDH. (I) The ubiquitination of overexpressed Vps55-GFP. Overexpressed Vps55-GFP was immunoprecipitated from yeast cells under denaturing conditions, and the IP products were analyzed by immunoblotting using antibodies against GFP and ubiquitin. (J) Vps55-GFP-Ub localization in ESCRT mutants. (K) Model of Vps55 recycling and degradation at the endosome. For all quantification shown in this figure, n = >30 cells from three independent experiments. Scale bar: 1 µm.