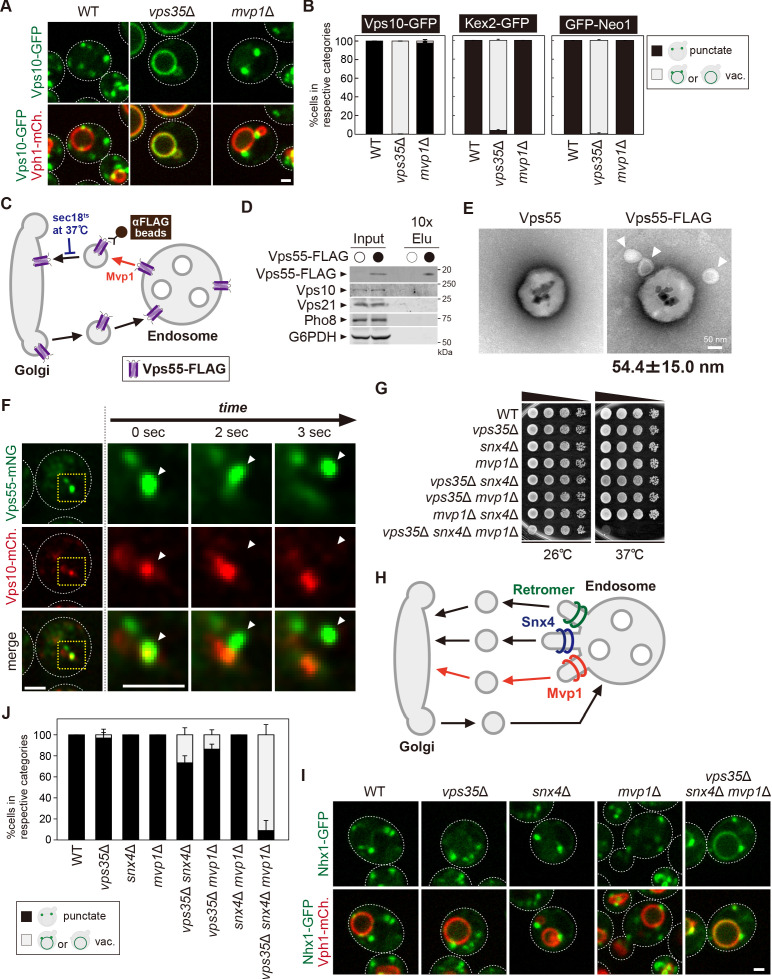
Figure 5. Mvp1 mainly mediates retromer-independent endosomal recycling.
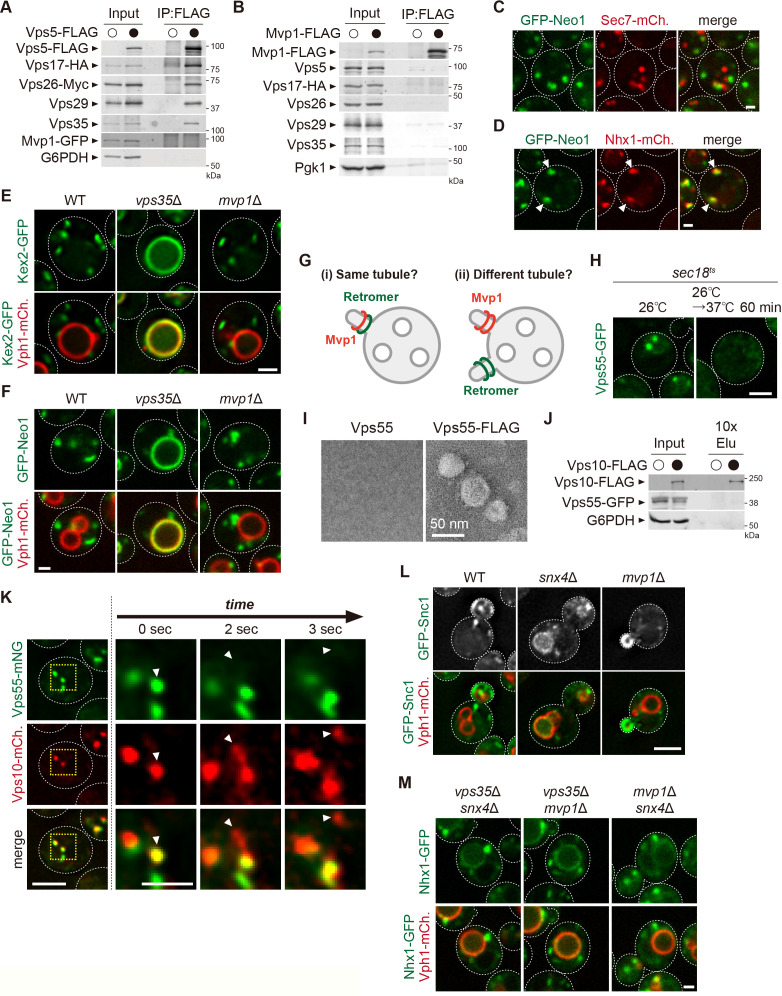
(A) Localization of Vps10-GFP in wild-type (WT), vps35Δ, and mvp1Δ cells. (B) Quantification of Vps10-GFP, Kex2-GFP, and GFP-Neo1 localization from A and Figure 5—figure supplement 1E,F, respectively. N = >30 cells from three independent experiments. (C) Schematic for immunoisolation of Vps55-FLAG-containing structures. (D) The immunoisolation of Vps55-containing vesicles. Vps55-FLAG-containing structures were immunoisolated from sec18ts mutants incubated at 37°C for 1 hr, and the isolated structures were analyzed by immunblotting using antibodies against FLAG, Vps10 (retromer cargo), Vps21 (endosome), Pho8 (vacuole), and glucose-6-phosphate dehydrogenase (G6PDH) (cytoplasm). (E) Electron microscopy (EM) analysis of the isolated Vps55-FLAG-containing structures from D. (F) Live-cell imaging analysis of Vps55-mNeonGreen and Vps10-mCherry. (G) Cell growth in vps35Δ snx4Δ mvp1Δ triple mutants. Cells lacking Vps35 as well as Snx4 and Mvp1 were grown at 26°C and 37°C. (H) Model of retromer-, Snx4-, and Mvp1-mediated recycling. (I) Nhx1 localization in SNX-BAR mutants. (J) Quantitation of Nhx1-GFP localization, from I and Figure 5—figure supplement 1M. N = >30 cells from three independent experiments. Scale bar: 1 µm.