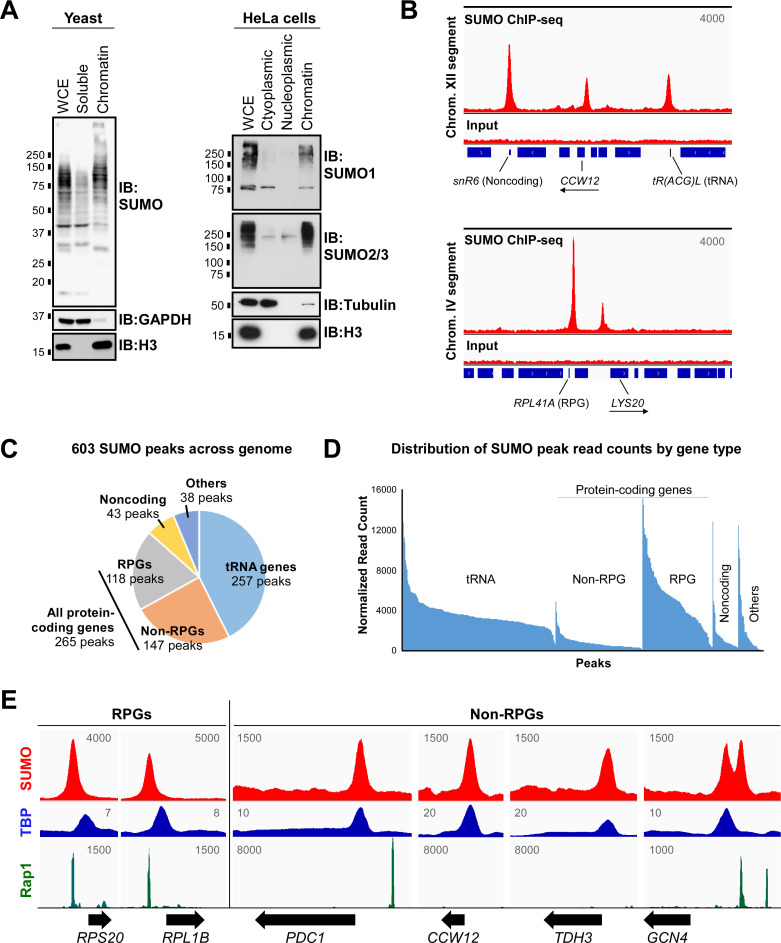
Fig 1. Sumoylated proteins associate stably with chromatin, including at many non-RPGs.
(A) Most sumoylated proteins are associated with chromatin. Fractionation experiments were carried out in normally growing, unmodified yeast and human HeLa cells, then whole cell extracts (WCE) and indicated fractions were analyzed by immunoblot (IB) with indicated antibodies, including antibodies for the yeast Smt3 peptide (“SUMO”), human SUMO1 or SUMO2/3 isoforms, and histone H3. (B) Independent duplicate SUMO ChIP-seq experiments were performed in yeast, and sample alignments are shown from Replicate 1, using the Integrative Genomics Viewer (IGV) genomic alignment tool, with reads from the SUMO IP and corresponding inputs over selected short segments of Chromosomes XII and IV. Values correspond to maximum data range (read numbers) for the view shown and blue bars along the bottom represent gene positions, including for two non-RPGs, CCW12 and LYS20, whose ORF orientations are indicated with arrows. See S9 Table for a list of yeast genes associated with the 603 SUMO peaks. (C) Pie chart showing fractions of the 603 SUMO ChIP-seq peak set associated with different gene types. Non-RPG refers to protein-coding genes that are not ribosomal protein genes (RPGs). See S2 Table for detailed description of peak classifications. (D) Distribution of read counts for the 603 SUMO ChIP-seq peaks, separated by gene type, then ranked by normalized read counts. Normalized read counts were determined using DiffBind tool with two independent ChIP-seq replicates. (E) Sample SUMO peak alignments from Replicate 1 are shown for two RPGs and four non-RPGs. Peak alignments for TBP and Rap1 are also shown, using published ChIP-seq and ChIP-exo datasets, respectively, for comparison with SUMO peak positions (NCBI GEO database accession numbers GSM2870615 and GSE93662, respectively). Values refer to maximum data range (read numbers) for the view shown, and unnormalized alignments were generated using IGV.