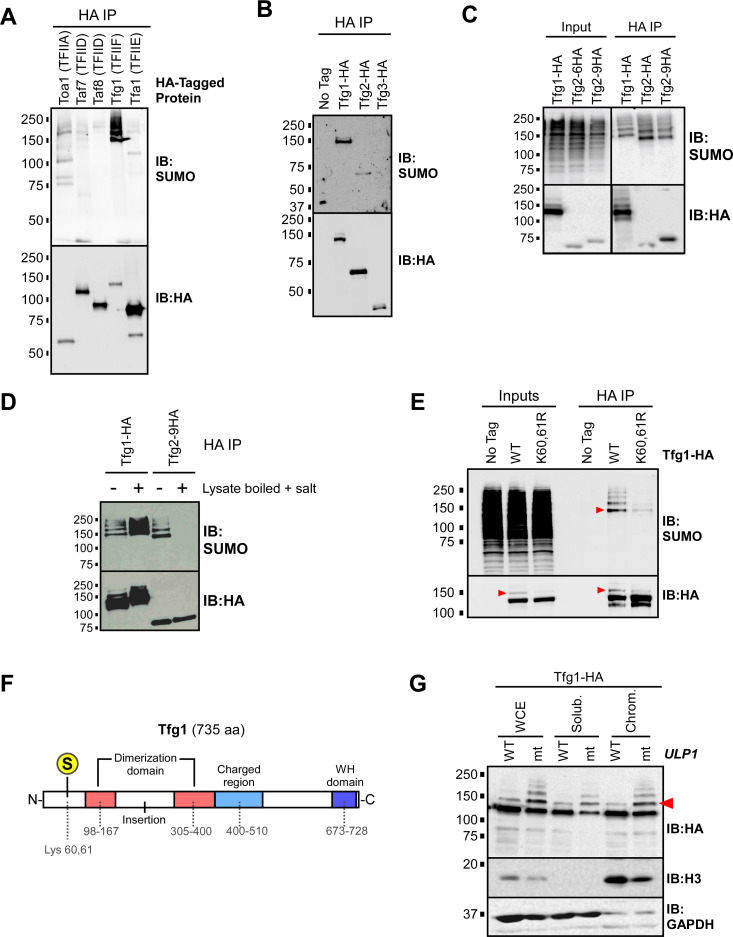
Fig 4. The large subunit of TFIIF, Tfg1, is sumoylated at Lys 60/61.
(A) Yeast strains were generated that each express a 6xHA C-terminal epitope tag on a different GTF subunit, including Toa1 (TFIIA), Taf7 (TFIID), Taf8 (TFIID), Tfg1 (TFIIF), or Tfa1 (TFIIE). Cultures of the strains were used to prepare lysates, under non-denaturing conditions, that were then used in HA IP experiments, followed by SUMO and HA immunoblots (IBs). (B) Strains expressing HA-tagged forms of TFIIF subunits Tfg1, Tfg2, or Tfg3 (also known as TAF14), were used for HA-IP experiments followed by SUMO and HA immunoblot analysis. “No tag” refers to the parental strain (W303a) that expresses no HA-tagged proteins. (C) Strains expressing Tfg1-HA, Tfg2-HA, or Tfg2-9HA (with a 9xHA tag instead of the usual 6xHA tag) were used in an HA IP analysis performed with lysates generated under non-denaturing conditions, followed by HA and SUMO immunoblots. The pattern detected in the SUMO blot of the Tfg2 IPs likely corresponds to sumoylated Tfg1 which coIPs with both forms of Tfg2. In the Tfg1-HA IP lane, the sumoylated species migrate slower because sumoylated Tfg1-HA includes the 6xHA tag, whereas Tfg1 (and its sumoylated forms) is untagged in the Tfg2-6HA and Tfg2-9HA strains. (D) Lysates were prepared from Tfg1-HA and Tfg2-9HA strains, which were then either treated (+) or not treated (-) by boiling for 5 min and adjusting the NaCl concentration to 0.5 M to promote the disruption of protein complexes prior to IP. Lysates were then used for HA IP analysis followed by SUMO and HA immunoblots. The pattern seen in the SUMO blot of the Tfg2-9HA IP disappears in the treated sample, implying that these sumoylated species are indeed derived from coIPed, sumoylated Tfg1. Note that treatment appears to elevate Tfg1 sumoylation levels (compare first two lanes in SUMO blot), likely because it inactivates naturally occurring SUMO proteases present in the lysate. (E) A mutant strain was generated that expresses Tfg1-HA with Arg substitutions at Lys 60 and 61 (K60,61R). This strain, along the WT Tfg1-HA-expressing strain, were used in HA IP-immunoblot experiments, and SUMO and HA immunoblots are shown. To disrupt protein-protein interactions, NaCl concentration was increased to 0.5 M and lysates were then boiled for 5 min, then cooled on ice prior to IP. Inputs represent approximately 5% of the material used for IP. (F) Diagram of Tfg1 domain structure, based on [64]. The major SUMO acceptor site is indicated with an encircled S at Lys 60/61. (G) Sumoylated Tfg1 associates with chromatin. A strain harboring a point mutation (I615N) in ULP1 (mt), and an isogenic wild-type strain (WT), were engineered to express Tfg1-HA, then the strains were used for chromatin fractionation analysis. Whole-cell extract (WCE), soluble, and chromatin fractions were analyzed by HA, histone H3, and GAPDH immunoblots. Red arrowheads indicate the position of mono-sumoylated Tfg1 in HA and SUMO immunoblots throughout.