Figure 3.
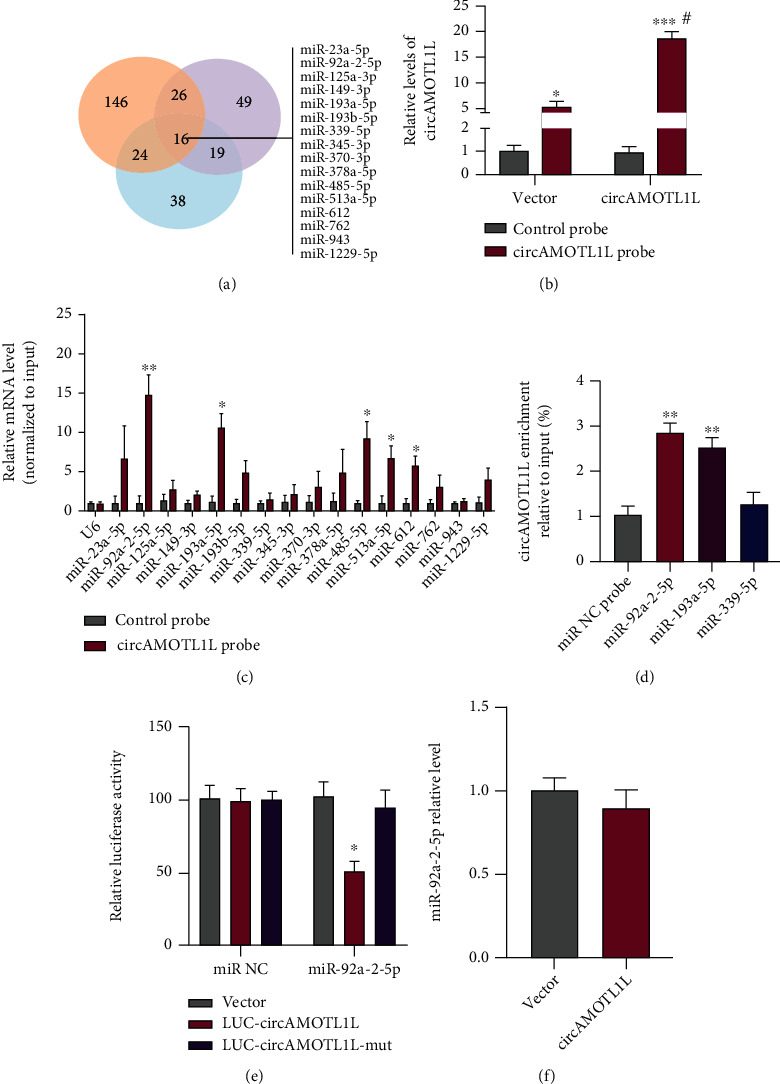
circAMOTL1L sponges miR-92a-2-5p in the RCC cell. (a) Venn diagram hybrid of target miRNA from three prediction programs (miRanda, RNAhybrid, and RNA22). (b) pcDNA-circAMOTL1L or empty vector was transfected into 786-O cells. qRT-PCR examined the pull-down efficiency of circAMOTL1L or NC probe. ∗p < 0.05, ∗∗∗p < 0.001, ##p < 0.01 vs. corresponding control. (c) qRT-PCR examined the relative expression of indicated miRNAs from pull-down precipitates. Normalized to control U6. ∗p < 0.05, ∗∗p < 0.01 vs. control probe. (d) qRT-PCR examined circAMOTL1L enrichment sedimented by biotin-labeled miR-92a-2-5p, miR-193a-5p, or miR-339-5p or NC probes. ∗∗p < 0.01 vs. NC probe. (e) Luciferase reporter assays examined luciferase activity of LUC-circAMOTL1L or LUC-circAMOTL1L-mut reporter constructs in 786-O cells cotransfected with the miR-92a-2-5p mimic or NC miR. ∗p < 0.05 vs. empty vector. (f) qRT-PCR examined miR-92a-2-5p expression in 786-O cells transfected with pcDNA-circAMOTL1L or empty vector. Graph bars represent mean ± SEM of three independent experiments.
