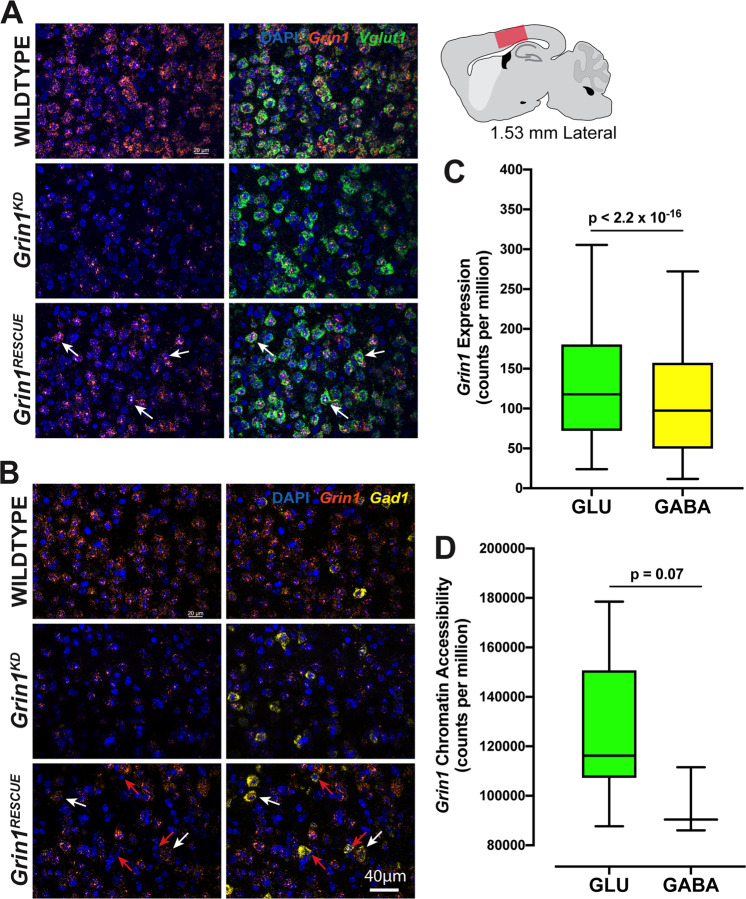
Fig. 2. Grin1 gene expression levels and chromatin accessibility of the adult mouse somatosensory cortex.
Grin1 mRNA expression in Vglut+ (a) and Gad1+ (b) cells in the adult mouse somatosensory cortex (1.53 mm lateral from midline). Grin1 (orange), Vglut1 (green) and Gad1 (yellow) mRNA was visualised in mouse sagittal sections (20 µm) with fluorescent in situ hybridisation in WT, Grin1KD, and Grin1RESCUE mice. Solid white arrows indicate cells with Grin1 expression, red arrows indicate cells without Grin1 expression. c Grin1 gene expression levels and d Grin1 chromatin accessibility in wild-type adult mouse visual cortex glutamatergic (Vglut1+) and GABAergic (Gad1+) cells. Data are from publicly-accessible single-cell transcriptomics data [16] and pooled cell type-specific ATAC-seq data [18] provided by the Allen Institute for Brain Science. Transcriptomic data are quantified as counts per million reads sequenced (CPM) and are based on exonic reads only. ATAC-seq data are quantified as counts per million nucleotides in locus. To aid visualisation, only cells up to the 95th percentile of Grin1 expression or coverage are shown. Data shown as box and whisker plots, 5–95 percentile, Wilcoxon rank-sum test between GLU and GABA.