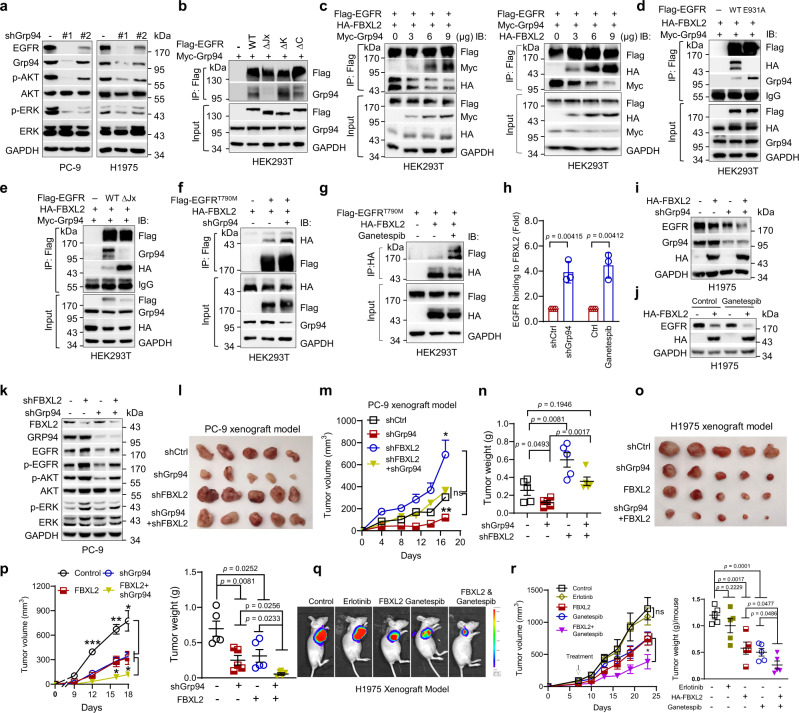
Fig. 5. Grp94 competes with FBXL2 for EGFR binding, and inhibition of Grp94 promotes FBXL2-mediated EGFR degradation in the suppression of TKI-resistant lung tumor growth.
a PC-9 or H1975 cells stably expressing shGrp94 were subjected to Western blot analyses. b, c HEK293T cells were co-transfected with indicated expressing plasmids for 36 h. Cells were then treated with MG132 for 4 h, followed by IP-Western analyses. d, e HEK293T cells were co-transfected with HA-FBXL2, Myc-Grp94, and indicated Flag-EGFR expressing plasmids for 36 h. Cells were then treated with MG132 for 4 h, followed by IP-Western analyses. HEK293T cells were co-transfected with Flag-EGFRT790M, HA-FBXL2, and either shCtrl or shGrp94 expressing plasmids overnight (f); or HEK293T cells were co-transfected with Flag-EGFRT790M and HA-FBXL2 expressing plasmids overnight and were then treated with or without ganetespib (8 nM) for 12 h (g). Cells were treated with MG132 for 4 h prior to IP-Western analyses. The affinity of EGFR binding to FBXL2 was quantified and presented as means ± SD. i H1975 cells stably expressing HA-FBXL2 or shGrp94, or both were subjected to Western blot analyses. j H1975 cells stably expressing HA-FBXL2 were treated with or without ganetespib (8 nM) for 24 h prior to Western blot analyses. PC-9 cells stably expressing either shFBXL2 or shGrp94, or both were subjected to k Western blot analyses or l−n xenograft tumor growth assays (n = 5/group). Photos of tumors, growth curve, and tumor weight were presented. o, p H1975 cells stably expressing either HA-FBXL2 or shGrp94, or both were subjected to xenograft tumor growth assays (n = 5/group). Photos of tumors, growth curve, and tumor weight were presented. q, r H1975 cells (5 × 105) stably expressing HA-FBXL2 or vector were subjected to xenograft tumor growth assays (n = 5/group). Mice were treated with erlotinib or ganetespib. Representative bioluminescence imaging, growth curve and tumor weight were presented. The experiment was repeated three times independently with similar results (a−g and i−k). Data were presented as means ± SEM (m, n, p, and r). *p < 0.05, **p < 0.01, ***p < 0.001; all by two-tailed Student’s t-test. Source data are provided as a Source data file.