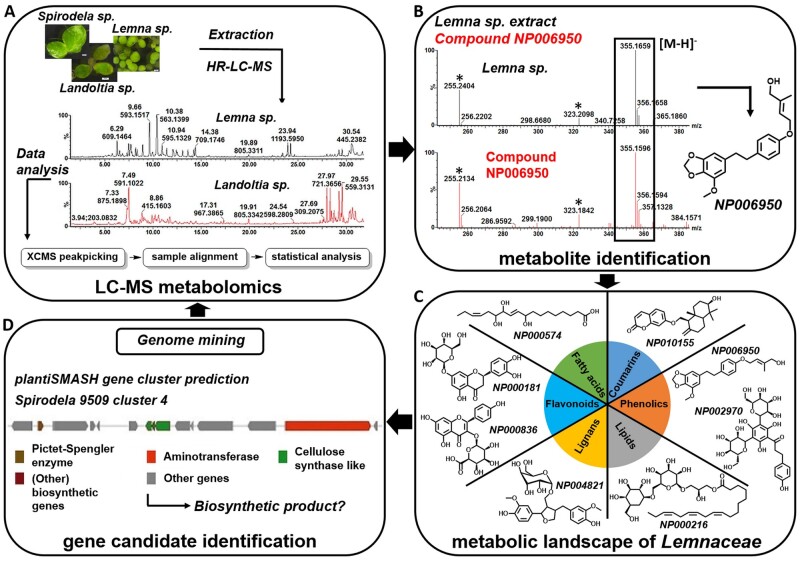
Figure 4.
Integration of metabolomics and genomics analysis in the Lemnaceae. (A) Comprehensive mass spectrometry-based metabolomics as a tool for pathway elucidation and characterization of biosynthetic enzymes and genes. Plant extracts are analyzed using high-resolution LC–MS (HR-LC–MS). Raw chromatograms typically consist of several thousand mass signals, and data can be deconvoluted using computation tools. (B) One method for compound identification from complex mixtures involves matching the mass spectra from an extract corresponding to a peak onto an entry in spectral libraries. An example for metabolite NP006950 (Weizmass library) is shown, illustrating that chromatographic retention, accurate mass, and mass fragmentation of a peak fraction in a Lemna gibba extract (upper black peak) matched a library entry (lower red peak) that can be assigned with high confidence (Shahaf et al., 2016). (C) These techniques, in combination with chemical classification, allow the metabolic landscape of Lemnaceae to be defined. (D) This knowledge can help identify metabolic genes/enzymes using either structure-based reaction prediction or genome mining approaches (such as metabolic gene cluster prediction that hints at possible biosynthetic pathways) as well as possible secondary metabolites (such as by correlation with the metabolomics dataset).