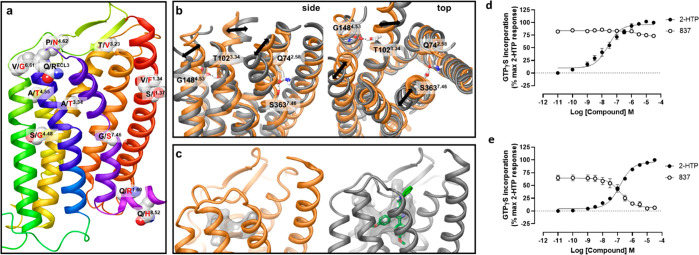
Figure 6.
Dynamics of mouse and human GPR84 homology models helps to predict amino residues responsible for species selectivity of the 1,2,4-triazine antagonists. Nonconserved residues between mouse and human orthologues are shown on the human GPR84 homology model in space-filling representation (a). The receptor model is shown in cartoon representation. Nonconserved residues of the third intracellular loop are omitted. Amino acids from human and mouse are shown in black and red and residue labels respectively using Ballesteros–Weinstein residue location numbering. Overlay of the average mouse (orange) and human (gray) GPR84 structures from MD simulations shown from side and top (from the receptor extracellular opening) views (b). Nonconserved residues and their counterparts in interhelical hydrogen bonding are shown in stick-like representation. Hydrogen bonds are shown as black dashed lines. Black arrows show the difference in the position of helices 3, 5, and 7. The orthosteric binding cavity of mouse (orange) and human (gray) GPR84 detected by the MDpocket program22 from MD simulation trajectories shown in transparent surface representation (c). Human GPR84 is shown bound to compound 837 (green). Ala102Thr-Gly363Ser human GPR84-Gαi2 and Thr102Ala-Ser363Gly mouse GPR84-Gi2α fusion proteins were produced and expressed stably in Flp-In TREx 293 cells (d, e). [35S]GTPγS binding assays performed on membranes of these cells showed that, although compound 837 was unable to effectively block activation of Ala102Thr-Gly363Ser human GPR84 by 2-HTP (d), it did effectively and fully block activation of Thr102Ala-Ser363Gly mouse GPR84 (e).