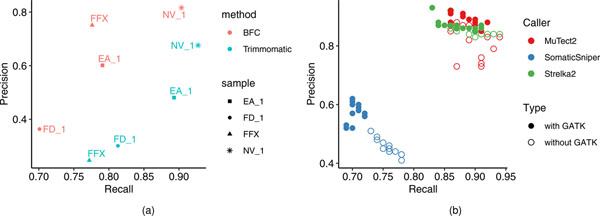
Extended Data Fig. 6 |. Effect of post alignment processing on precision and recall of WES and WGS run on FFPE DNA.
(a) precision and recall of mutation calls by Strelka2 on BWA alignments. A single library of FFPE DNA (FFX) and three libraries of fresh DNA (EA_1, FD_1, and NV_1) were run on a WES platform. Resulting reads were either processed by the BFC tool or by Trimmomatic. processed FASTQ files were then aligned by BWA and called by Strelka2. precision and recall were derived by matching calling results with the truth set. (b) precision and recall of mutation calls by three callers, Mutect2 (blue), Strelka2 (green), and SomaticSniper (red), on BWA alignments without or with GATK post alignment process (indel realignment & BQSR).