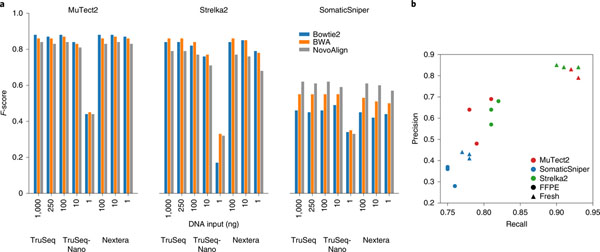
Fig. 3 |. Nonanalytical factors affecting mutation calling.
a, Caller performance on three library preparation protocols with different DNA Inputs: 1, 10, 100, 250 and 1,000 ng. WGS sequencing on TruSeq and TruSeq-Nano libraries was performed at LL, while WGS sequencing on Nextera libraries was performed at IL. All sequencing experiments were performed with HiSeq 4000 and analyzed using three aligners (BWA, Bowtie2 and NovoAlign) and three callers (MuTect2, Strelka2 and SomaticSniper). b, Performance of MuTect2, Strelka2 and SomaticSniper on WGS with fresh DNA or FFPE DNA (24 h) from a BWA alignment.