Figure 3.
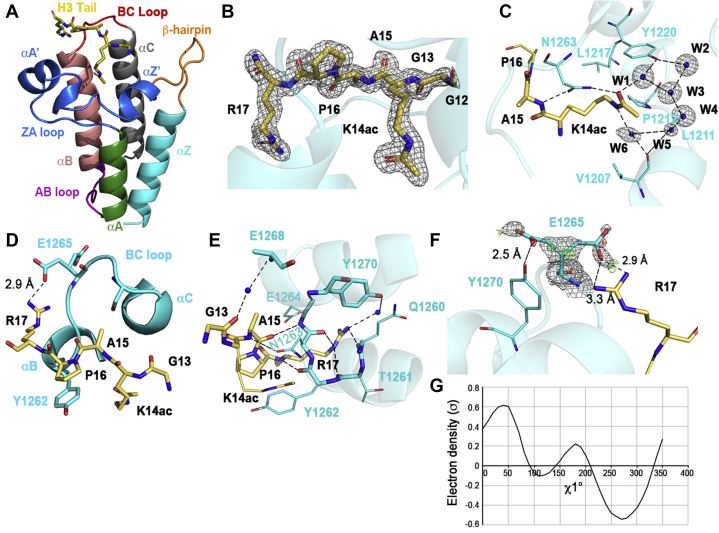
Structure of the CeSMARCA4 BD in complex with its modified histone H3K14ac target.A, structural features of the overall 1.29 Å CeSMARCA4 bromodomain–H37–20K14ac complex. The αZ, αA, αB, and αC helices are colored cyan, green, dark salmon, and gray, respectively. The ZA, AB, and BC interhelical loops are colored blue, purple, and red, respectively. The ZA loop includes two short helices, αA′ and αZ′, with a 310 helical turn adjacent to αA’. The β-hairpin, a hallmark of family VIII bromodomains, is shown in orange. The modified histone H3 peptide is shown in yellow. B, residues G13-R17 can be unequivocally traced in the 2Fo-Fc electron density map, which is contoured at 1σ, while G12 can be partially traced. C, interactions with K14ac (yellow, stick) bound in the CeSMARCA4 binding pocket (cyan). The conserved Asn1263 side chain forms a hydrogen bond with the acetyl carbonyl group of acetyl-lysine and also forms a hydrogen bond with the H3A15 amide nitrogen atom. A network of structural water molecules (blue spheres in 2Fo-Fc electron density map contoured at 1σ) mediate interactions between K14ac and the Val1207 carbonyl group, and Tyr1220 side chain. Hydrophobic interactions with the side chains of Val1207, Leu1211, Pro1212, and Leu1217 further stabilize K14ac in the bromodomain pocket. D, sidechain interactions of the H3 tail outside of the K14ac binding pocket. H3A15 binds in a small pocket adjacent to the BC loop. H3P16 contributes hydrogen stacking interactions, packing with the benzene ring of Tyr1262. And H3R17 forms a salt bridge with one conformation of the Glu1265 sidechain. E, backbone interactions by the bromodomain with the H3 tail outside of the K14ac binding pocket. Glu1264 and Tyr1262 form direct backbone contacts with the carbonyl of A15, and amide nitrogen of R17. The bromodomain Glu1268 amide nitrogen forms a water-mediated contact with the G13 carbonyl group. Three additional hydrogen bonds mediate R17 sidechain interactions: by Gln1260 and Thr1261 backbone contacts and a water-mediated contact with Tyr1270. F, the two conformations of Glu1265. The dominant rotamer (60% occupancy) forms a salt bridge with H3R17. The other rotamer (40% occupancy) forms a hydrogen bond with Tyr1275. The final 2mFo-DFc electron density (grey) is shown contoured at 0.8 σ. Very little +3σ (green) or −3σ (red) mFo-DFc density remain in the map. Maps were generated with Phenix (67) and displayed in Pymol (69). G, ringer plot of Glu1265 shows two chi1 peaks at ∼60° and ∼180° (40).