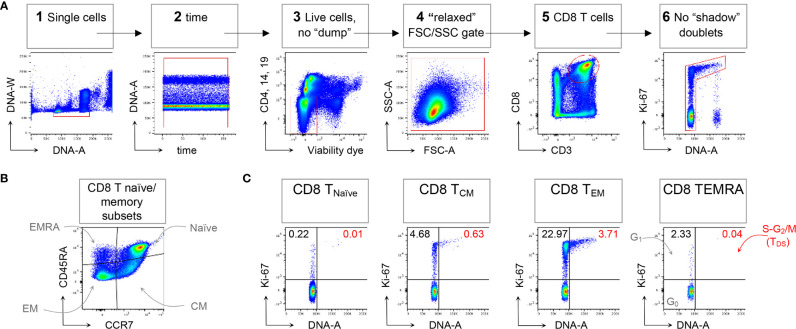
Figure 1.
Example of CD8 T cell naïve/memory subset analysis by TDS assay. HD PBMCs were stained with the viability dye eFluor 780 (eF780), the DNA dye Hoechst-33342, and fluorochrome conjugated mAbs against surface markers and Ki-67, as described (16). An example of flow cytometry analysis is shown. (A) Gating of viable single CD8 T cells in 6 steps: 1) DNA-A/-W singlets. Single cells having 2n≤ DNA content ≤4n were selected on the DNA-area (A) versus (vs) DNA-width (W) plot; 2) Time exclusion. Stable acquisition over time (seconds) was monitored on the time vs DNA-A plot and any events collected in case of pressure fluctuations were excluded; 3) Viable cells, no “dump”. Cells expressing CD4, CD14 and CD19, and dead cells were excluded; 4) FSC A/SSC-A “relaxed” gate. A “relaxed gate was used on the FSC-A vs SSC-A plot, to include highly activated and cycling lymphocytes (15); 5) CD8 T cells. CD8 T cells were gated on the CD3 versus CD8 plot; 6) Refined singlets. A few remaining doublets composed by one cell sitting on top of another (so called “shadow” doublets) were excluded as Ki-67int/- events having > 2n DNA content (16). This gating strategy was used as a base for the subsequent gates. (B) The following naïve/memory subsets of CD8 T cells were identified: CD45RA+ CCR + Naïve, CD45RA- CCR7+ central memory (CM), CD45RA- CCR7- effector memory (EM), and CD45RA+ CCR7- (EMRA). (C) Cell cycle phases of each naïve/memory CD8 T cell subset were defined on DNA-A vs Ki67-A plot as follows: cells in G0 were identified as DNA 2n/Ki67- (bottom left quadrant); cells in G1 as DNA 2n/Ki67+ (upper left quadrant); cells in S-G2/M (or TDS cells) as DNA>2n/Ki67+ (top right quadrant). Unpublished data in relation to (16).