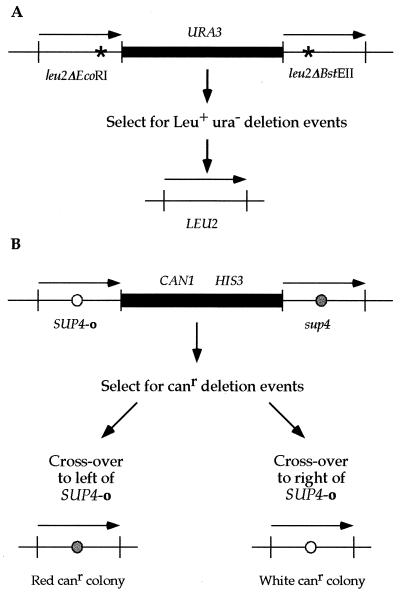
FIG. 1.
The direct repeat recombination constructs for leu2 and SUP4 are depicted. Both assays utilize a direct repeat of two alleles that are separated by plasmid and selectable marker sequences. Recombination events between the repeats that result in the retention of one allele and deletion of the intervening sequences can be selected. The leu2 and SUP4 direct repeats are 2.4 kb and are not drawn to scale. (A) In the leu2 assay (46), both alleles contain a frameshift mutation created by filling in a restriction enzyme site (EcoRI or BstEII). Leu+ recombinants are first selected for on medium lacking leucine and further identified as Ura− colonies after replica plating on medium lacking uracil. (B) The SUP4 assay (24) was modified by disrupting URA3 with HIS3 as described in Materials and Methods. The alleles differ by a single nucleotide change in the anticodon. Canr recombinants are selected on canavanine-containing medium. The strain contains the ochre suppressible ade2-1 allele, which enables colony color to designate which SUP4 allele is retained in the genome after direct repeat recombination: sup4 colonies are red, while SUP4-o colonies remain white. Deletions are confirmed by their failure to grow on histidine-less medium.