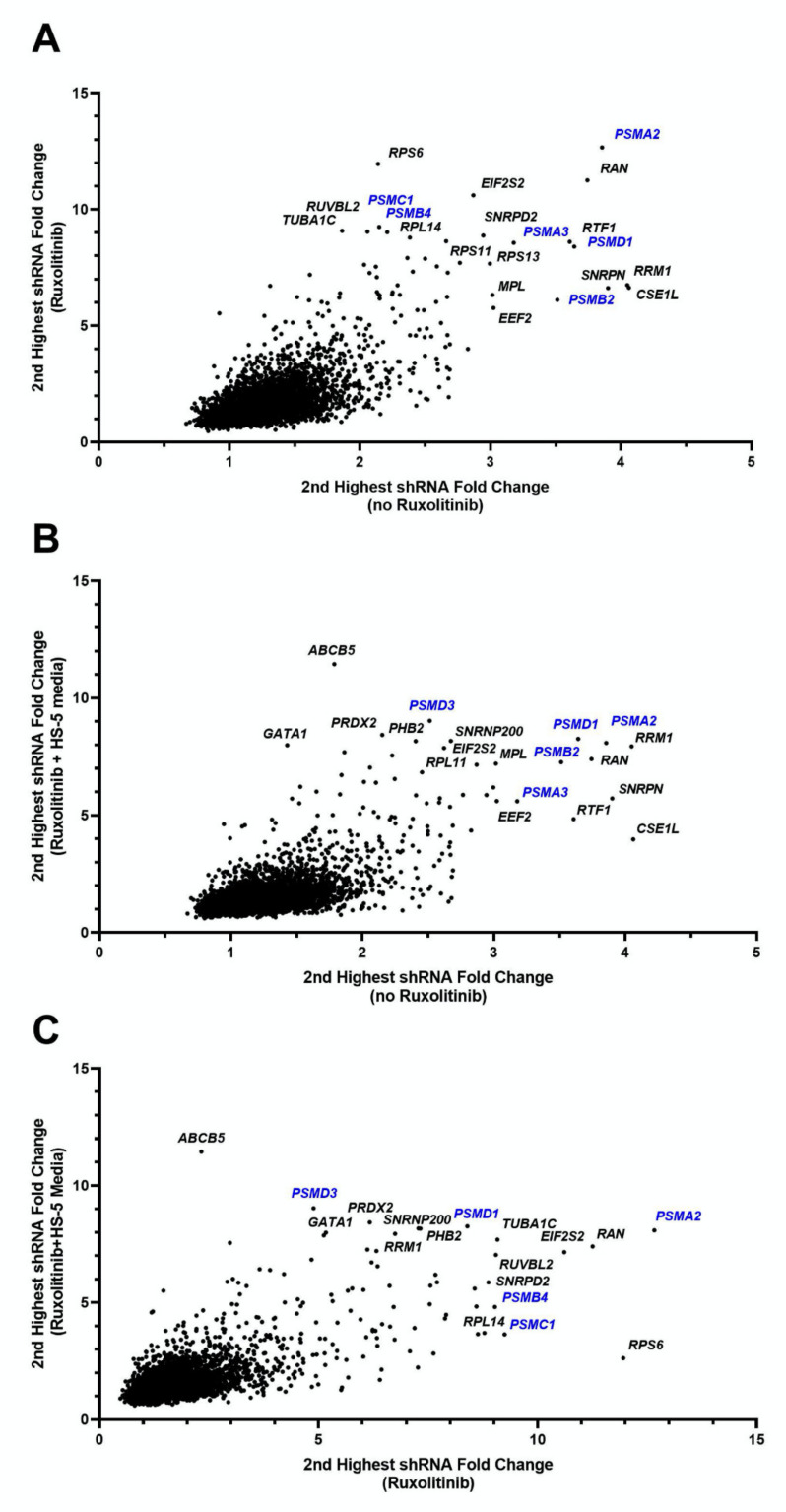
Figure 2.
Fold change of shRNAs for HEL cell line under various conditions. Each figure shows the second highest fold change for shRNAs targeting each of 4974 genes, showing comparisons for each combination of conditions. Genes in the uppermost right corners indicate genes which under both of the compared conditions had high shRNA depletion at follow-up compared to baseline, indicating essentiality of the gene for HEL cells survival. (A) Fold change of ruxolitinib treated cells to baseline (vertical axis) versus cells cultured without ruxolitinib to baseline (horizontal axis). (B) Fold change of HS-5 media plus ruxolitinib treated cells to baseline (vertical axis) versus cells cultured without ruxolitinib to baseline (horizontal axis). (C) Fold change of HS-5 media plus ruxolitinib treated cells to baseline (vertical axis) versus cells cultured with ruxolitinib (and no HS-5 media) to baseline (horizontal axis). The first replicate (run 1) values are shown for each setting. These data indicate a significant abundance of proteasome genes (in blue font), requisite for the survival of HEL cells regardless of microenvironment or exposure to ruxolitinib.