ABSTRACT
The type 4 secretion system (T4SS) represents a bacterial nanomachine capable of trans-cell wall transportation of proteins and DNA and has attracted intense interest due to its roles in the pathogenesis of infectious diseases. In the current investigation, we uncovered three distinct gene clusters in Clostridioides difficile strain 630 encoding proteins structurally related to components of the VirB4/D4 type 4C secretion system from Streptococcus suis strain 05ZYH33 and located within sequences of conjugative transposons (CTn). Phylogenic analysis revealed that VirB4- and VirD4-like proteins of the CTn4 locus, on the one hand, and those of the CTn2 and CTn5 loci, on the other hand, fit into separate clades, suggesting specific roles of identified secretion system variants in the physiology of C. difficile. Our further study on VirB4- and VirD4-like products encoded by CTn4 revealed that both proteins possess Mg2+-dependent ATPase activity, form oligomers (most likely hexamers) in aqueous solutions, and rely on potassium but not sodium ions for the highest catalytic rate. VirD4 binds nonspecifically to DNA and RNA. The DNA-binding activity of VirD4 strongly decreased with the W241A variant. Mutations in the nucleotide sequences encoding presumable Walker A and Walker B motifs decreased the stability of the oligomers and significantly but not completely attenuated the enzymatic activity of VirB4. In VirD4, substitutions of amino acid residues in the peptides reminiscent of Walker structural motifs neither attenuated the enzymatic activity of the protein nor influenced the oligomerization state of the ATPase.
IMPORTANCE C. difficile is a Gram-positive, anaerobic, spore-forming bacterium that causes life-threatening colitis in humans. Major virulence factors of the microorganism include the toxins TcdA, TcdB, and CDT. However, other bacterial products, including a type 4C secretion system, have been hypothesized to contribute to the pathogenesis of the infection and are considered possible virulence factors of C. difficile. In the current paper, we describe the structural organization of putative T4SS machinery in C. difficile and characterize its VirB4- and VirD4-like components. Our studies, in addition to its significance for basic science, can potentially aid the development of antivirulence drugs suitable for the treatment of C. difficile infection.
KEYWORDS: ATPase, Clostridioides difficile, VirB4, VirD4, secretion systems
INTRODUCTION
Clostridioides (formerly Clostridium) difficile is a Gram-positive, anaerobic, spore-forming bacterium causing C. difficile infections (CDIs) in humans, which include antibiotic-associated diarrhea, colitis, and pseudomembranous colitis (1). Studies on the molecular pathogenesis of CDI demonstrated that the ability of the bacterium to cause disease depends largely on the production of toxins—two large glucosylating toxins (TcdA and TcdB) and the binary ADP-ribosylating toxin CDT (2–6). Deeper knowledge of the pathogenesis of CDI suggests that other microbial products in addition to toxins can be of high value for the pathogenicity of the infection agent. Such nontoxin virulence factors can include surface structures of the bacterium participating in motility and adhesion (7–13).
Recently, components of a type 4 secretion system (T4SS) have been found in C. difficile genomes within the sequences of conjugative transposon 2 (CTn2), CTn4, and CTn5 (14–16). The biological functionality of T4SS has not been directly demonstrated in C. difficile, although transposons containing T4SS components were shown to be excised from the genome of strain 630 and transferred to strain CD37 (17).
In general, the T4SS represents a nanomachine capable of the transportation of proteins and DNA into eukaryotic target cells, horizontal interbacterial transfer of mobile genetic elements, and exchange of DNA with the outer space (18). Functionally, T4SSs can be subdivided into two major classes. One class participates in the interbacterial transfer of DNA through conjugation or by exporting/importing DNA to/from the extracellular space. This results in horizontal transfer of mobile genetic elements that are beneficial for microbial population traits. Another class is found in Gram-negative bacteria, such as Agrobacterium tumefaciens and Legionella, is directly important for the pathogenesis of the corresponding diseases, and accomplishes its functions through the delivery of effector DNA or proteins into target eukaryotic cells, thereby manipulating a plethora of eukaryotic functions (19, 20).
On a structural basis, the T4SS has been classified into types A, B, and C. The type representatives of T4SS-A include the A. tumefaciens VirB/VirD4 T4SS and Escherichia coli conjugation machinery of the R388 and pKM101 plasmids. In the former case, T4SS-A is composed of 12 subunits, each in multiple copies, termed VirB1 through VirB11 and VirD4: (i) the cytoplasmic ATPases (VirB4, VirB11, and VirD4), (ii) components of an inner membrane complex (VirB3, VirB6, and VirB8), (iii) components of an outer membrane complex (VirB7, VirB9, and VirB10), and (iv) pilus components (VirB1, VirB2, and VirB5). The prototype T4SS-B is the Dot/Icm secretion system of Legionella pneumophila. It is composed of 27 subunits, of which only a few are structurally related to those of T4SS-A while being functionally similar (18, 21, 22).
Recently, a new variant of T4SS (T4SS-C) was identified within the 89-kb pathogenicity island of Streptococcus suis (23). Only four components of this secretion system (VirB1, VirB4, VirB6, and VirD4) appeared to be sufficient for its function. As suggested by the authors, a VirB1-like sequence encodes an amidohydrolase with the CHAP (cysteine, histidine-dependent amidohydrolase/peptidase) domain, which is responsible for punching holes in the peptidoglycan; VirB6 probably forms the transportation channel across the cell wall, and VirB4 and VirD4 proteins energize the whole secretion device (24). Interestingly, intact T4SS-C is necessary for the development of streptococcal toxic shock syndrome (23, 25, 26) and secretion of subtilisin-like proteinase and the potential effector molecule SP1 (24, 27, 28).
Since components of T4SS occurring in pathogenic C. difficile isolates might play roles in the pathogenesis of CDI (14, 15), we aimed to characterize VirB4- and VirD4-like proteins, elements of this newly identified secretion system.
RESULTS
Characterization of VirB4/D4-coding loci in the C. difficile 630 genome.
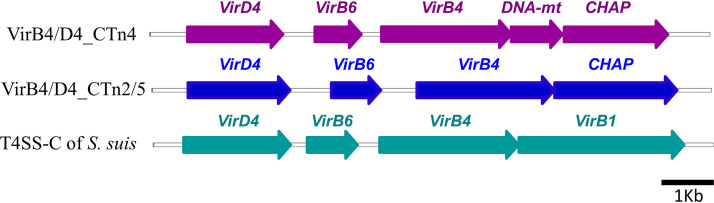
As a first step toward the characterization of the VirD4 and VirB4 proteins, we looked into the organization of T4SS loci in the genome of C. difficile 630, utilizing the data on T4SS-C of S. suis 05ZYH33 (24). Using amino acid sequences of VirB4 (NCBI database protein tag ABP89935.1) and VirD4 (ABP89939.1) of S. suis in a BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi), three loci containing similar proteins could be identified in C. difficile (Fig. 1). Since the loci were found within the nucleotide sequences of CTn2, CTn4, and CTn5, they were named VirB4/D4_CTn2, VirB4/D4_CTn4, and VirB4/D4_CTn5, respectively.
FIG 1.
Genetic organization of VirB4/VirD4 loci in C. difficile 630. DNA-mt, DNA methyltransferase; CHAP, CHAP domain-containing protein. The coding sequences are approximately to scale. See Table S1 for precise identification of the C. difficile 630 T4SS components.
The three VirB4/D4 loci consist of VirD4-, VirB6-, and VirB4-coding genes in an orientation identical to that in S. suis. The last coding sequence in the S. suis operon is virB1. In contrast, C. difficile contains an unstudied open reading frame in this location, annotated as “CHAP domain-containing protein.” In the case of VirB4/D4_CTn4, there is an additional DNA methyltransferase-coding sequence between the genes coding for VirB4 and CHAP domain-containing protein. VirB4 and VirD4 from the CTn4 locus were 56 to 58% and 74% identical to the corresponding proteins encoded by the CTn2 and CTn5 loci, respectively. The identity of the matching proteins within the CTn2 and CTn5 loci exceeded 90%; therefore, they were collectively considered CTn2/5. VirB4 and VirD4 of S. suis contained 31 to 36% amino acid residues identical to VirB4_CTn2/4/5 or VirD4_CTn2/4/5 (see Fig. S1 in the supplemental material). These data suggested that according to the primary sequences, the identified C. difficile proteins represented components of two types of putative secretion machineries, VirB4/D4_CTn4 and VirB4/D4_CTn2/5, each of which was only distantly related to the described previously canonical T4SS-C apparatus from S. suis (24).
To obtain more information on the relationship of C. difficile T4SS variants with known secretion systems, we performed BLAST searches utilizing VirB4 and VirD4 protein sequences from CTn4 and CTn2/5 clusters as queries. Surprisingly, the database examinations revealed several groups of bacterial products that were over 90% identical to VirB4 and VirD4 from C. difficile and clearly different from the components of previously investigated T4SS-C from S. suis or E. faecalis (Fig. S2). Thus, the CTn2/5 group contained VirB4/D4 components from pathogenic Gram-positive microorganisms such as enterococci, streptococci, and peptostreptococci, while the CTn4 group included VirB4/D4 proteins from organisms of the poorly studied genera Blautia, Coprococcus, and Eisenbergiella. Since we failed to find any data on the T4SS organization of the latter bacteria, we focused our further investigations on the VirB4 and VirD4 components of CTn4.
Purification of recombinant VirB4_CTn4 and VirD4_CTn4 proteins.
According to the nucleotide sequence data, VirD4_CTn4 (WP_011861117.1) is a 595-amino-acid protein with a molecular mass of 68 kDa, while VirB4_CTn4 (WP_011861114.1) consists of 809 amino acid residues with a calculated mass of 91 kDa.
The coding sequences were amplified using PCR and were then cloned into a pET28a vector. However, our attempts to produce full-size proteins in E. coli strains transformed with the corresponding plasmids were unsuccessful. As seen in SDS-polyacrylamide gels, only minute amounts of the proteins could be isolated by HisTrap affinity chromatography, and the quality of the purified proteins was not sufficient for further studies (Fig. S3). Bearing in mind that the investigated proteins are products of a Gram-positive microorganism, we tried a commercially available Bacillus megaterium expression system for the production of VirD4 and VirB4. In contrast to VirD4, VirB4 could be isolated from B. megaterium in sufficiently pure form. However, the purified protein lacked any ATPase activity (Fig. S4).
VirB4 and VirD4 consist of several structural components, including the COOH-terminal nucleotide-binding domain (NBD), which facilitates ATPase activity (29). Due to our special interest in the enzymatic properties of the VirB4/D4 proteins, we performed NH2-terminal truncations by PCR and cloned the coding sequences of the remaining COOH-terminal NBDs into pET28 vectors. The engineered delVirB4 and delVirD4 consisted of amino acid residues 438 to 809 and 110 to 595, respectively, fused to a 6×His-containing NH2-terminal tag. Unfortunately, as with the full-size proteins, production of the deleted versions of the proteins in E. coli was also insufficient for biochemical analysis (data not shown).
To improve the expression levels of the delVirB4 and delVirD4 proteins, we utilized the maltose-binding protein (MBP) fusion vector pMal-c5x. The addition of the normally well-produced hydrophilic MBP tag to the protein sequences considerably increased the expression levels of the coding sequences and kept the bacterial products in soluble form, thus helping to isolate relatively large amounts of the enzymes (Fig. S3B). Purification of delVirB4 and delVirD4 from liquid Terrific broth (TB) cultures resulted in a higher yield than that from LB, although the proteins appeared more degraded. This fact eliminated any advantages of the high protein production in the former medium. Thus, using pMal-based plasmids, we were able to repeatedly isolate 3 to 4 mg of pure protein from 1 liter of liquid LB culture. Therefore, in subsequent experiments, deletion-containing versions of VirB4 and VirD4 were used as MBP-tagged constructs.
Enzymatic characterization of delVirB4_CTn4 and delVirD4_CTn4.
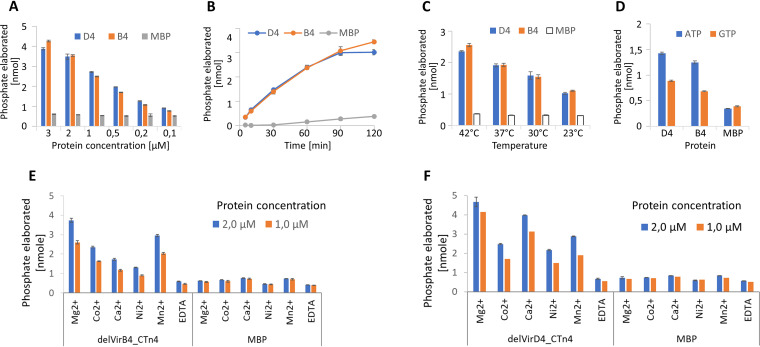
Initial experiments with different concentrations of delVirB4 and delVirD4 demonstrated dose–response kinetics. In contrast to a negative control (MBP), increasing concentrations of C. difficile proteins resulted in the efficient hydrolysis of added ATP over a 1-h period at 35°C (Fig. 2A; Fig. S5A). Time curve data demonstrated an almost linear phosphate release curve over a 2-h period for delVirB4, while delVirD4 appeared to be less stable at later time points under the experimental conditions (Fig. 2B). Both proteins were active at temperatures of 23°C to 42°C (Fig. 2C; Fig. S5B). A higher temperature appeared to be detrimental for delVirD4, while delVirB4 tolerated incubation at 42°C for 30 min quite well. Last, the two proteins demonstrated an obvious preference for ATP over GTP (Fig. 2D).
FIG 2.
Influence of multiple parameters on NTPase activity of delVirD4_CTn4 (D4) and delVirB4_CTn4 (B4). (A) Proteins at the indicated concentrations were incubated at 35°C with 100 μM ATP for 1 h. (B) Proteins at 1 μM were incubated with 100 μM ATP for the indicated times. (C) Proteins at 1 μM were incubated with 100 μM ATP at indicated temperatures for 30 min. (D) Proteins at 1 μM were incubated at 35°C for 1h with 50 μM ATP or GTP. (E and F) The indicated cations (as chloride salts) or EDTA (each at 2 mM) was added to the reaction mixtures, containing 100 μM ATP and 1 μM or 2 μM delVirB4_CTn4 (E) or delVirD4_CTn4 (F), and incubated at 35°C for 1 h.
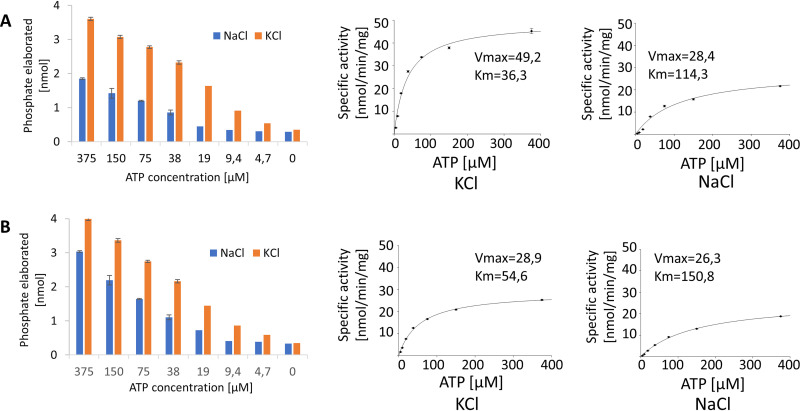
In the next set of experiments, we studied the influence of cations on the enzymatic performance of delVirB4 and delVirD4. The addition of EDTA completely blocked the reaction. The highest ATPase activity was seen with MgCl2 for both proteins, followed by MnCl2 and CaCl2 with delVirB4 and delVirD4, respectively (Fig. 2E and F). When potassium was used as a substitute for sodium ions in a reaction mix, it significantly stimulated the ATPase activity of both proteins, as clearly seen from the kinetic data (Fig. 3).
FIG 3.
Dependance of ATPase activity on potassium versus sodium cation. DelVirB4_CTn4 (A) and delVirD4_CTn4 (B) at 0.25 μM were incubated for 1 h at 35°C with different concentrations of ATP in Tris-HCl buffer containing 2 mM MgCl2 and 150 mM NaCl or 75 mM KCl.
Oligomer formation by delVirB4_CTn4 and delVirD4_CTn4.
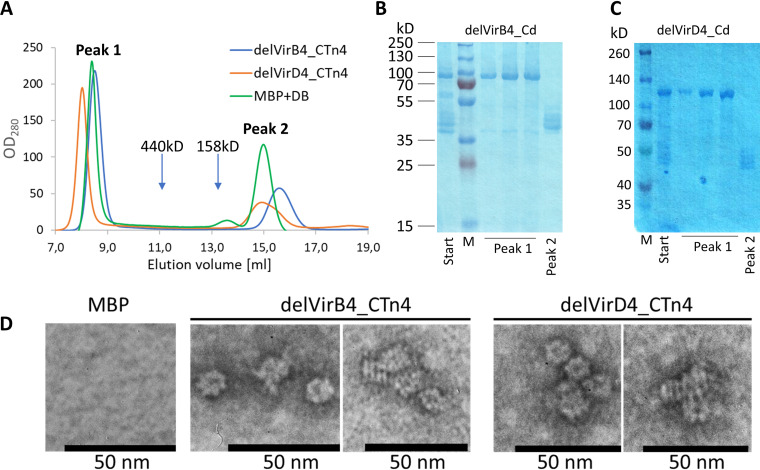
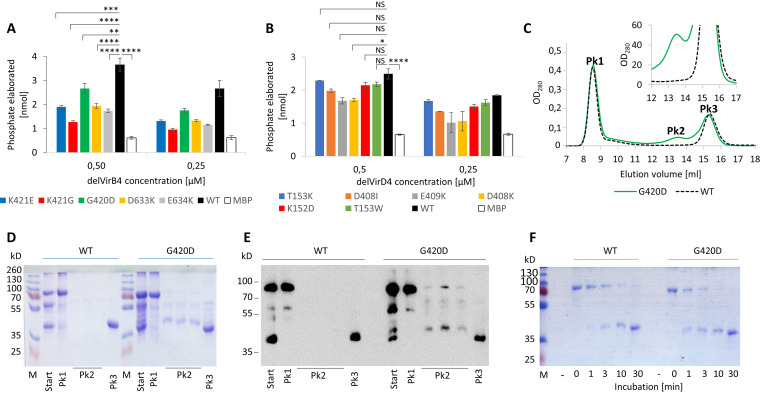
To investigate the stoichiometry of delVirB4 and delVirD4 under native conditions, we used gel chromatography. Concentrated preparations of the ATPases were loaded onto a Superdex200 column and eluted with TBS. Surprisingly, no protein peaks were seen at the expected elution volumes (∼14 ml for proteins ∼100 kDa). Instead, two peaks could be observed at elution volumes of ∼8 ml (which is close to the void volume of the column) and approximately 15 ml (where proteins of 40 to 50 kDa should elute) (Fig. 4A). SDS-PAGE analysis showed that the material that eluted near the void volume contained delVirB4 and delVirD4, while fragments of 40 to 50 kDa could be seen in 15 ml of material (Fig. 4B and C). These data indicated that in aqueous solutions, both proteins formed high-molecular-mass oligomers. Bearing in mind the technical characteristics of the Superdex200 10/30 column, the molecular mass of the protein complexes should exceed 600 kDa, suggesting oligomer formation with a stoichiometry of ≥6 monomers.
FIG 4.
Oligomerization of MBP-tagged delVirB4_CTn4 and delVirD4_CTn4. (A) Overlay of partial Superdex200 chromatograms with delVirB4_CTn4, delVirD4_CTn4, and a mixture of MBP (45 kDa) plus dextran blue (DB; 2,000 kDa) as size markers. Approximate positions of ferritin (440 kDa) and aldolase (158 kDa) are indicated by the arrows. (B and C) SDS-PAGE analysis of MBP-tagged delVirB4_CTn4 (B), and delVirD4_CTn4 (C) loaded on (start) and eluted from (peaks 1 and 2) Superdex200 columns. Lane M, molecular mass markers. (E) Proteins after MBPtrap chromatography were additionally purified by Superdex200 (see analysis of peak 1 fractions in panels B and C), diluted to 10 μg/ml with distilled water (dH2O), and subjected to negative staining/transmission electron microscopy. Samples were processed as described in Materials and Methods. Bar, 50 nm.
To further study oligomerization processes of the delVirB4 and delVirD4 proteins, we used electron microscopy with negative staining. During these experiments, we observed particles of ∼10 nm, predominantly demonstrating two types of silhouettes. Most of them were circular and exhibited 6- to 7-fold symmetry, while others had a striated rectangular look (Fig. 4D). Such structural variability probably resulted from different plane positions during sample processing in electron microscopy.
Structure-function analysis of delVirB4_CTn4 and delVirD4_CTn4.
To probe the structure-function relationship in delVirB4 and delVirD4, we mutated the amino acid residues within the regions known to be important for the catalytic activity of the ATPases. In relation to delVirB4, these substituted residues included G420 and K421, belonging to a Walker A motif, and D633 and E634 of Walker B. With delVirD4, the substituted residues were K152 and T153 of Walker A and D408 and E409 in Walker B motifs (Fig. S1). In accordance with the literature (30), all engineered mutations decreased the ATPase activities of the VirB4 protein. Interestingly, increasing the negative net charge in the Walker A motif of delVirB4 by introducing D420 or E421 produced less of an effect than changing the side chain structure in the G421 variant. Mutations in the Walker B-coding sequence (D633K and E634K), resulting in inversion of charge in the corresponding sites, appeared to suppress the ATPase activities of delVirB4 more prominently than mutations of Walker A (Fig. 5A and B). Mutagenesis experiments performed on VirD4 failed to demonstrate any involvement of the substituted amino acid residues in catalysis.
FIG 5.
ATPase activity and oligomer stability of site-substituted delVirB4_CTn4 and delVirD4_CTn4. (A and B) The indicated amino acid residues were substituted by QuikChange mutagenesis on the corresponding wild-type coding sequences. Enzymatic reaction was performed with two protein concentrations (0.5 and 0.25 μM) and 150 μM ATP. Results shown are means from 2 independent experiments with 3 measurement replicates each. Error bars represent standard deviations (n = 6). Student's t test was applied for statistical comparisons. *, P < 0.1; **, P < 0.05; ***, P < 0.001; ****, P < 0.0005; NS, not significant. (C to E) Oligomer stability of the wild type and G420D delVirB4_CTn4 variant. delVirB4_CTn4 variants purified by MBP tag chromatography were subjected to Superdex200 chromatography in 20 mM Tris-HCl (pH 7.4) plus 75 mM KCl (C). Thereafter, starting material (Start) and that of the major peaks (Pk 1, 2, and 3) were analyzed by 10% SDS-PAGE (D) and Western blotting with anti-MBP serum (E). Fractions of peak 1 obtained from the wild type (WT) and the G420D VirB4 variant were pooled and subjected to analytical trypsinolysis (1 ng of trypsin per 2 μg of protein) for 1, 3, 10, and 30 min at 25°C and SDS-PAGE analysis (F).
In the next set of experiments, we compared the oligomer structure stabilities of the wild-type delVirB4/D4 proteins with those of the site mutants. Using Superdex200 gel chromatography, SDS-PAGE, Western blotting, and trypsin cleavage experiments, we demonstrated that all amino acid substitutions in delVirB4 but not in delVirD4 variants resulted in oligomer instability, manifested by the appearance of an additional peak on the chromatograms (Fig. 5C; Fig. S6). This peak contained monomeric full-size delVirB4 and its fragments (Fig. 5D and E). Although the majority of the molecules remained in an oligomeric state and constituted the material of peak 1, trypsinization experiments verified its higher susceptibility to cleavage in the tested G420D variant in contrast to the wild-type ATPase (Fig. 5F).
Nucleic acid-binding activity of delVirB4_CTn4 and delVirD4_CTn4.
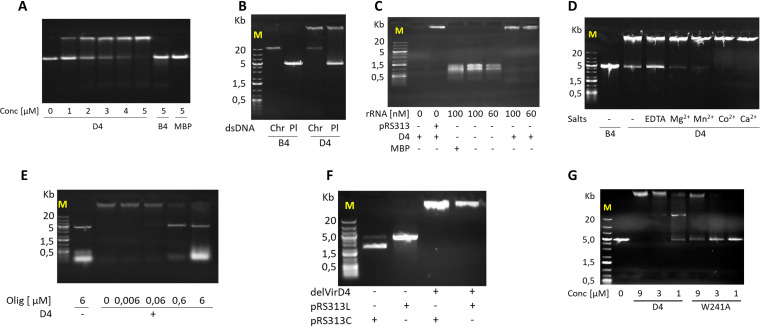
In the next set of experiments, we studied the interactions of C. difficile ATPases with DNA and RNA. To this end, following incubation on ice for 30 min, mixtures of delVirB4 or delVirD4 with plasmid double-stranded DNA (dsDNA), chromosomal dsDNA, or rRNA were separated by 0.5% agarose gel electrophoresis. In contrast to delVirD4, delVirB4 did not bind dsDNA. Conversely, delVirD4 demonstrated stable dsDNA-protein and rRNA-protein interactions, illustrated by the gel shifts of ethidium bromide-stained DNA and RNA fragments (Fig. 6). Chelation of cations by EDTA significantly decreased the DNA-binding activity of delVirD4, while the addition of divalent salts restored complex formation (Fig. 6D; Fig. S7). To test whether ssDNA is also able to interact with delVirD4, we performed competition experiments in which the preformed protein-plasmid complex was treated with oligonucleotide single-stranded DNA (ssDNA). The addition of the 81-mer oligonucleotide primer in increasing amounts displaced dsDNA from the preformed complex, which was evidenced by the appearance of an ∼5-kb plasmid band (Fig. 6E). With the plasmid, complex formation was independent of the form (linear or circular) of the added DNA molecule (Fig. 6F). Site substitution of W241A in a presumed nucleic acid-binding region (31) resulted in a strong decrease in DNA binding by delVirD4 (Fig. 6G). Interestingly, the mutation influenced neither the ATPase activity of the protein nor its oligomerization state (Fig. S6B and S8).
FIG 6.
Interaction of delVirB4_CTn4 (B4), wild-type delVirD4_CTn4 (D4), and W241A variant of delVirD4 (W241A) with nucleic acids. (A) Binding of plasmid dsDNA (pRS313; 3.3 nM) with the C. difficile proteins at different concentration. (B) Binding of the C. difficile proteins (10 μM) with plasmid (Pl; 3.3 nM) and chromosomal (Chr; 50 ng) dsDNA. (C) Binding of the delVirD4_CTn4 protein (D4) with rRNA. RNA at the indicated concentrations (or pRS313 at 3.3 nM as a positive control) was mixed with 10 μM delVirD4_CTn4 (or MBP) and processed as described in Materials and Methods. (D) Binding of the C. difficile proteins (3 μM) with plasmid dsDNA (pRS313; 3.3 nM) in the presence of added EDTA or metals salts (2 mM each). Addition of EDTA or metal cations to plasmid DNA without C. difficile proteins did not change nucleic acid mobility patterns (Fig. S6). (E) Interaction of delVirD4_CTn4 with synthetic DNA oligonucleotide ssDNA. delVirD4_CTn4 (10 μM) was incubated with pRS313 (2 nM) for 10 min on ice. Thereafter, the indicated amounts of oligonucleotide ssDNA were added, and incubation on ice continued for 30 min. (F) Plasmid pRS313 was linearized by BamHI/EcoRI cleavage or left untreated as circular. Thereafter, linear (pRS313L) and circular (pRS313C) plasmid dsDNA (3.3 nM each) were mixed with delVirD4_CTn4 C. difficile protein (10 μM) and incubated for 10 min on ice. Agarose gel electrophoresis was performed in 0.5% (A, B, C, D, and F) and 1.5% (E) agarose. (G) Influence of mutations on interaction of delVirD4_CTn4 with plasmid DNA. Wild-type and mutated delVirD4_CTn4 at the indicated concentrations was incubated with the plasmid pRS313 (2.5 nM) for 10 min on ice. Thereafter, the samples were subjected to electrophoresis in 0.5% agarose. M, nucleic acid marker. Sizes of major fragments are shown on the left, in kilobases.
Because the binding of DNA can affect the enzymatic properties of C. difficile proteins, we studied the influence of DNA on the ATPase activity of delVirB4 and delVirD4. This was not obviously the case, given that the addition of pRS313 (dsDNA) or synthetic oligonucleotide (ssDNA) to the ATPase reaction mix did not change the enzymatic activity of the proteins (Fig. S9).
DISCUSSION
Nine types of microbial secretion systems are known to date, among which type 4, comprising type 4A and type 4B subfamilies, is relatively well studied and is able to deliver its substrates (DNA or proteins) to eukaryotic or prokaryotic cells (19, 32). Recently, subfamily T4SS-C was found in C. difficile (14, 15), although neither its biological functionality nor the biochemical properties of its components have been studied.
Genes encoding VirD4, VirB4, and VirB6 can be identified in C. difficile 630. However, instead of the VirB1 coding sequence present in S. suis, a CHAP domain-containing protein is encoded in the C. difficile chromosome as the last gene of the operon. CHAP is generally involved in partial degradation of the cell walls of Gram-positive microorganisms during cell division or phage release (33, 34). Moreover, a CHAP-related sequence can be found as a domain in a VirB1-like protein of S. suis (24). Based on these data, one can speculate that the coding sequence, termed the CHAP domain-containing protein, can accomplish the functions of VirB1 in C. difficile and is involved in local degradation of the clostridial cell wall during T4SS channel assembly.
An interesting feature of the VirB4/D4_CTn4 operon is the presence of a DNA methyltransferase coding sequence. There are three known major forms of DNA methylation in prokaryotes, accomplished by DNA methyltransferases: 6-methyladenine, 4-methylcytosine, and 5-methylcytosine. Increasing evidence suggests that DNA methylation regulates a number of biological processes of high medical importance, including antibiotic resistance, virulence, evasion of the host immune response, and replication niche adaptation (35–38). In a recent paper, conserved 6-methyladenine methyltransferase (camA) was identified as an important factor for sporulation of C. difficile and the pathogenesis of experimental CDI (39). Whether a DNA methyltransferase encoded by the VirB4/D4_CTn4 locus is involved in the pathogenicity of C. difficile is unknown, as is the relationship of the DNA methyltransferase with the T4SS-C function.
The major components of T4SSs include two VirB4- and VirD4-related ATPases. As seen from our protein homology searches, VirB4_CTn4 and VirB4_CTn2/5 fall into two different clades, as do VirD4_CTn4 and VirD4_CTn2/5, which, in turn, are distinct from the clade containing the previously described “classical” T4SS-C of S. suis (24). Intriguingly, some strains of S. suis possess an operon related to VirB4/D4_CTn2/5 of C. difficile instead of “classical” T4SS-C. These data illuminate a fruitful direction of further research on the structure-function variability in the type 4 secretion machinery of Gram-positive microorganisms.
Most biochemical data on VirB4 and VirD4 were obtained by studying proteins originating from Gram-negative microorganisms, predominantly from E. coli (40–43). Bearing in mind the low sequence similarity of the clostridial ATPases with those of E. coli T4SS, it was interesting to study the catalytic properties of VirB4 and VirD4 homologs from the Gram-positive human pathogen C. difficile. We found that both delVirB4_CTn4 and delVirD4_CTn4 preferred higher temperatures for enzymatic activity, up to 42°C. This was in some contrast to previous data on the E. coli VirD4 homolog TrwB that demonstrated an optimal temperature range of 33 to 37°C, with a drastic decrease in activity at temperatures of 42°C or higher (31). Similar to TraE and TraK (VirB4 and VirD4 homologs in Aeromonas veronii T4SS, respectively), the best cofactor in the reaction with the clostridial proteins was Mg2+. However, the second most efficient cations with delVirB4_CTn4 and delVirD4_CTn4 were Mn2+ and Ca2+, while for TraE and TraK, these were Mn2+ and Co2+ (44). In line with previous data on T4SS ATPases, sodium ions were strongly inhibitory for C. difficile VirB4/D4 enzymes and should be replaced in the reaction by potassium (40).
Several studies with purified VirB4/TraB, VirB4/TrwK, and VirD4/TrwB of E. coli demonstrated that the proteins in water solution behaved as enzymatically active hexamers (40, 42, 43, 45). These data were confirmed by gel filtration, sedimentation velocity analysis, native polyacrylamide gel electrophoresis, and electron microscopy. However, in other experiments, purified VirB4 behaved as a monomer (29, 30). In our experiments, both delVirB4_CTn4 and delVirD4_CTn4 formed high-molecular-mass oligomers and hexameric particles, as seen in gel chromatography and electron microscopy, respectively. Moreover, as observed during Superdex200 chromatography, the oligomerization rates for both proteins were close to 100%, since no traces of monomers or dimers could be found on the chromatograms.
Two peptides are known to represent general nucleotide-binding motifs of ATP-requiring enzymes: Walker A (or P loop) and Walker B. The consensus sequence of Walker A was originally identified as GXXXXGKT/SXXXXXXI/V, while Walker B conserved features are represented by R/KXXXGXXXLhhhDE (“h” stands for a hydrophobic amino acid residue) (46). According to the crystal structure of the VirD4/TrwB hexamer, Walker A and Walker B are located on the surface of each oligomer on the interface between the neighboring subunits. Walker A is preceded by a β-strand, followed by an α-helix, and forms a loop around bound ATP. Amino acid residues of the motif interact with phosphates of the nucleotide and Mg2+ (47). It is thus not surprising that amino acid substitutions in Walker A resulted in drastic decreases in both ATPase and conjugation-stimulating activities (31, 48). Putative Walker A and Walker B motifs are easily detected in the VirB4_CTn4 sequence. We performed site-specific mutagenesis experiments and engineered a panel of amino acid substitutions to study their influence on the ATPase activity of the proteins. All engineered proteins demonstrated significantly lower but still detectable levels of enzymatic activity, suggesting the presence of cryptic catalytic centers in the proteins. Indeed, several conserved sequences marking ATP-processing proteins are known. In the case of VirB4, these include regions C (DX[D/E]XXXE), D (RK), and E (S/TQ) (29). Additionally, a cryptic but functional novel ATP-binding site has been identified in the N-terminal domain of VirB4 (42). We did not perform any site-directed mutation experiments on these zones of C. difficile ATPases. Interestingly, attenuation of ATPase activity paralleled less efficient oligomer formation, as observed in our gel chromatography and trypsinization experiments. Therefore, we speculate that diminished enzymatic activities in the mutated VirB4 variants were caused primarily by structural perturbations in the molecules, resulting in less efficient formation of functionally competent oligomers.
In contrast to VirB4, Walker A and Walker B sequences are not easily seen within the VirD4_CTn4 sequence. While we identified peptides that can represent the motifs and performed a set of mutation experiments, we are not absolutely sure of the correct identification of Walker A and Walker B positions in this case, since the results of our enzymatic experiments were negative.
Since VirB4/D4 T4SSs are known to participate in transporting DNA, we were interested in whether VirB4 and VirD4 ATPases of C. difficile could bind nucleic acids. According to our gel shift experiments, delVirD4_CTn4 but not delVirB4_CTn4 formed complexes with DNA and RNA. In relation to VirD4, our results are in line with published data. Indeed, several VirD4-like proteins have been shown to interact with both dsDNA and ssDNA (48–51). With VirB4, the data are controversial. VirB4/TraB, the ATPase of the E. coli conjugation plasmid pKM101, formed complexes with DNA, as demonstrated by agarose gel shift and proteolysis protection assays (42), while VirB4/TrbE of the pRP4 plasmid and VirB4/TrwK of R388 did not (30). We are not aware of any published data showing the interaction of VirD4 ATPase with RNA. Our experiments did demonstrate this binding behavior. It is interesting to speculate whether RNA could be a T4SS substrate and whether RNA translocation is important in some way for the physiology or pathogenicity of microorganisms.
Previously, it was shown that replacement of W216, located in the central pore of the VirD4/TrwB E. coli hexamer, resulted in a protein that did not hydrolyze ATP and exerted a dominant negative effect on R388 conjugation (31). These tryptophan residues from adjacent monomers form a ring, which possibly participates in DNA binding and translocation. In our experiments, alanine substitution of the similarly located amino acid residue W241 produced a protein with wild-type ATPase activity and normal oligomerization properties but diminished DNA-binding activity.
In summary, we identified novel variants of T4SS-C in the important human pathogen C. difficile and characterized the biochemical properties of major VirB4- and VirD4-like ATPases. Our findings expand the list of bacterial secretion systems that are potentially important for bacterial physiology and pathogenicity and should be accompanied in the future by the demonstration of T4SS functionality in C. difficile.
MATERIALS AND METHODS
Materials and bacterial strains.
Restriction endonucleases, T4 DNA ligase, Phusion DNA polymerase, molecular mass markers, and kits for DNA isolation were from Thermo Fisher Scientific (Moscow, Russia). Lysogeny broth Miller recipe (LB) medium was from Amresco (Solon, OH, USA); Terrific broth (TB) medium, brain heart infusion, and yeast extract were from Difco (Franklin Lakes, NJ, USA). Liquid chromatography media were from GE Healthcare (Moscow, Russia), and reagents for agarose and polyacrylamide gel electrophoresis and general laboratory reagents were from Merck (Moscow, Russia).
Molecular cloning and recombinant protein production were performed in Escherichia coli DH10B, Rosetta(DE3) (Merck) and Bacillus megaterium WH320 (MoBiTec GmbH, Göttingen, Germany). Plasmids for cloning and recombinant protein expression in E. coli were based on pUC19 (New England Biolabs), pET28a-c (cloning with an NH2-terminal 6×His tag; Merck), and pMal-c5x (cloning with the NH2-terminal maltose-binding protein [MBP] tag; New England Biolabs). The vector pHis1522 (MoBiTec) was used for expression in B. megaterium. C. difficile 630 (TcdA+ TcdB+ CDT+ human isolate [16]) was used as a source of DNA for gene cloning. Anti-MBP antibody was purchased from New England Biolabs (number E8032S).
Gene cloning.
For cloning the VirD4_CTn4 and VirB4_CTn4 coding sequences, the corresponding genes were PCR amplified from the genomic DNA of C. difficile with the primers 808/809 and 812/813, respectively (primer information is presented in Table S2 in the supplemental material). The synthesized sequences were cut with NdeI and EcoRI restriction endonucleases and ligated into a similarly digested pET28a vector, producing plasmids p28-VirD4_CTn4 and p28-VirB4_CTn4. To make 5′-terminal truncations of VirD4_CTn4 and VirB4_CTn4 coding sequences, plasmids p28-VirD4_CTn4 and p28-VirB4_CTn4 were used in PCR as matrix DNA with the primers 1014/17 and 1038/17. The resulting amplified nucleotide sequences were digested with BamHI and EcoRI endonucleases and ligated into pET28b and pET28a to yield p28-delVirD4_CTn4 and p28-delVirB4_CTn4, respectively. For site-directed mutagenesis of putative Walker A and Walker B motifs, the gene fragment from p28-delVirB4_CTn4 was cloned into pUC19 using BamHI/EcoRI restriction endonuclease sites. Following the QuikChange reaction (Stratagene, Moscow, Russia) with the corresponding primers (Table S2), mutated sequences were reintroduced back into pET28. QuikChange mutagenesis of VirD4 was performed directly on the p28-delVirD4_CTn4 plasmid. In a few cases with VirD4, where the QuikChange protocol did not result in successful nucleotide substitutions, PCR splicing reactions were performed (52). To produce coding sequences with the additional MBP-containing tag, the plasmids p28-delVirD4_CTn4 and p28-delVirB4_CTn4 as well as their site-mutated variants were digested with NcoI/SalI, and the isolated insert sequences were ligated into pMal-C5x. The resulting plasmids were named pMal-delVirD4_CTn4 and pMal-delVirB4_CTn4 for the wild-type genes. Indications of the engineered amino acid substitution for the corresponding protein variants were added to the plasmid name where applicable.
To engineer plasmids for expression in B. megaterium, full-size VirB4_CTn4 and VirD4_CTn4 were amplified from p28-VirB4_CTn4 and p28-VirD4_CTn4 using primers 474/475 and ligated into SacI/KpnI restriction endonuclease sites of pHis1522, producing pHis1522-VirB4_CTn4 and pHis1522-VirD4_CTn4.
Recombinant-protein purification.
For recombinant-protein production, the E. coli Rosetta strain was grown in LB medium supplemented with the corresponding antibiotics (ampicillin at 100 μg/ml for pMal-c5x or kanamycin at 50 μg/ml for pET28 plasmids) until an optical density at 600 nm (OD600) of 0.5 was reached. Induction of expression was performed overnight with 0.5 mM isopropyl-β-d-1-thiogalactopyranoside (IPTG) at 22°C or for 1 h with 1 mM IPTG at 37°C with pET28 or pMal-c5X-based plasmids, respectively. For the production of proteins in Gram-positive hosts, recombinant B. megaterium was grown in 3.7% brain heart infusion–0.5% yeast extract liquid medium with tetracycline at 10 μg/ml to an OD600 of 0.3. Induction of expression was performed with 0.5% xylose overnight at 22°C. Both E. coli and B. megaterium were lysed by sonication. The resulting extracts were subjected to centrifugation at 30,000 × g for 30 min to separate cytosolic (water-soluble) and membrane (water-insoluble but 6 M urea-soluble) fractions. Recombinant proteins were subsequently purified via affinity chromatography using HisTrap or MBPtrap columns connected to an ÄKTA Explorer liquid chromatography system (GE Healthcare) according to the manufacturer’s instructions. The purified proteins were stored in 10% glycerol–20 mM Tris-HCl (pH 7.4)–150 mM NaCl (TBS) at −20°C.
Determination of ATPase activity.
The ATPase activity of the purified proteins was determined using two kits, an Enliten ATPase assay (FF2000; Promega, Moscow, Russia) and a malachite green phosphate assay (MAK307; Merck). Both assays were performed in two steps as detailed in the corresponding user manuals. Typically, the reaction mixture in the first step consisted of 0.1 mM ATP, a 1 μM concentration of the protein of interest, 2 mM MgCl2, 20 mM Tris-HCl (pH 7.4), and 150 mM NaCl in a total volume of 30 μl or 80 μl for the Enliten and malachite green assays, respectively. The reaction proceeded for 1 h at 35°C. Thereafter, in the second step, 30 μl of luciferin/luciferase reagent or 20 μl of malachite green reagent was added. The results were read on a GloMax-Multi+ plate reader immediately for the Enliten assay or following incubation at 22°C for 30 min for the malachite green assay. With the Enliten assay, the intensity of the emitted light following the addition of luciferin/luciferase reagent was proportional to the ATP concentration. Thus, measurement of the light intensity using a luminometer permits direct quantitation of nondegraded ATP present in the reaction mix. For the malachite green assay, the intensity of the developing green color is proportional to the concentration of phosphate, elaborated during ATP hydrolysis, and was measured at 600 nm. Here, to convert the optical density, displayed as optical units (OU600), to the amount of degraded ATP, a calibration curve was constructed by plotting the OUs obtained with phosphate standards against the amount of added phosphate.
Study of oligomer formation by delVirB4_CTn4 and delVirD4_CTn4.
Oligomerization of delVirB4_CTn4 and delVirD4_CTn4 proteins was studied by gel chromatography on a Superdex200 10/30 GL column connected to an ÄKTA Explorer liquid chromatography system (GE Healthcare). The proteins, dissolved in TBS at concentrations of ∼1.5 mg/ml, were loaded onto a column equilibrated in TBS or 20 mM Tris-HCl (pH 7.4)–75 mM KCl (TBK) and were eluted at 0.5 ml/min. Dextran blue, ferritin, aldolase, MBP, bovine serum albumin, and ovalbumin were used as molecular mass standards.
The ultrastructure of delVirB4_CTn4 and delVirD4_CTn4 oligomers was studied using a JEM 2100Plus negative-staining method with a JEM 2100Plus transmission electron microscope (JEOL, Japan). A drop of each purified protein at a concentration of 0.1 to 0.01 mg/ml in distilled water was placed on a Formvar copper grid (SPI Supplies, USA) for 60 s. The excess solution was removed using filter paper. The copper grids with the samples were stained with 1% uranyl acetate (Serva, Heidelberg, Germany) for 60 s, and the excess stain was then removed using filter paper. The grids were dried at room temperature for 10 min and analyzed using a transmission electron microscope at an accelerating voltage of 160 kV.
Determination of DNA/RNA-binding activity.
To determine dsDNA-binding activity, purified MBP-tagged proteins with NH2-terminal deletions at different concentrations were mixed with 50 ng of C. difficile chromosomal DNA or with 3.3 nM pRS313 plasmid DNA (53) linearized with BamHI/EcoRI restriction endonucleases. The mixtures were kept on ice for 10 min and then subjected to 0.5% agarose gel electrophoresis in 40 mM Tris-20 mM acetate-1 mM EDTA (TAE) buffer and stained with ethidium bromide. The interaction of ssDNA with delVirD4_CTn4 was studied in competition experiments, in which an initially formed dsDNA-protein complex of 2 nM pRS313 and 10 μM C. difficile protein was treated with increasing amounts (0.006 to 6 μM) of an 81-mer DNA oligonucleotide on ice for 30 min. Thereafter, the reaction was visualized by 1.5% agarose gel electrophoresis as described above. To study the interaction of RNA with VirD4_CTn4, we used RNA isolated from yeast ribosomes (a generous gift from Sabine Rospert, Freiburg University, Germany) (54) by an RNeasy kit (Thermo Fischer Scientific). Purified RNA at concentrations of 60 nM and 100 nM was added to the indicated proteins and processed as described above for dsDNA.
General biochemical methods.
Purified protein preparations were analyzed by polyacrylamide gel electrophoresis (PAGE) in sodium dodecyl sulfate (SDS)-containing buffer (55) and Western blotting with the anti-MBP antibody (56). The gels were stained with colloidal Coomassie G-250 (PageBlue; Thermo Fisher Scientific) or silver nitrate (57). Protein concentrations were estimated using Coomassie brilliant blue G-250 stain calibrated with bovine serum albumin as a standard (58).
ACKNOWLEDGMENT
The study was partially supported by the Russian Ministry of Public Health (project АААА-Н-18-118032390061-0).
Footnotes
Supplemental material is available online only.
Contributor Information
Yury Belyi, Email: belyi@gamaleya.org.
Conrad W. Mullineaux, Queen Mary University of London
REFERENCES
- 1.Abt MC, McKenney PT, Pamer EG. 2016. Clostridium difficile colitis: pathogenesis and host defence. Nat Rev Microbiol 14:609–620. 10.1038/nrmicro.2016.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aktories K, Papatheodorou P, Schwan C. 2018. Binary Clostridium difficile toxin (CDT)—a virulence factor disturbing the cytoskeleton. Anaerobe 53:21–29. 10.1016/j.anaerobe.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Aktories K, Schwan C, Jank T. 2017. Clostridium difficile toxin biology. Annu Rev Microbiol 71:281–307. 10.1146/annurev-micro-090816-093458. [DOI] [PubMed] [Google Scholar]
- 4.Chandrasekaran R, Lacy DB. 2017. The role of toxins in Clostridium difficile infection. FEMS Microbiol Rev 41:723–750. 10.1093/femsre/fux048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerding DN, Johnson S, Rupnik M, Aktories K. 2014. Clostridium difficile binary toxin CDT: mechanism, epidemiology, and potential clinical importance. Gut Microbes 5:15–27. 10.4161/gmic.26854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwan C, Kruppke AS, Nolke T, Schumacher L, Koch-Nolte F, Kudryashev M, Stahlberg H, Aktories K. 2014. Clostridium difficile toxin CDT hijacks microtubule organization and reroutes vesicle traffic to increase pathogen adherence. Proc Natl Acad Sci USA 111:2313–2318. 10.1073/pnas.1311589111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anjuwon-Foster BR, Maldonado-Vazquez N, Tamayo R. 2018. Characterization of flagellum and toxin phase variation in Clostridioides difficile ribotype 012 Isolates. J Bacteriol 200:e00056-18. 10.1128/JB.00056-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barketi-Klai A, Hoys S, Lambert-Bordes S, Collignon A, Kansau I. 2011. Role of fibronectin-binding protein A in Clostridium difficile intestinal colonization. J Med Microbiol 60:1155–1161. 10.1099/jmm.0.029553-0. [DOI] [PubMed] [Google Scholar]
- 9.Ðapa T, Dapa T, Leuzzi R, Ng YK, Baban ST, Adamo R, Kuehne SA, Scarselli M, Minton NP, Serruto D, Unnikrishnan M. 2013. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J Bacteriol 195:545–555. 10.1128/JB.01980-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacs-Simon A, Leuzzi R, Kasendra M, Minton N, Titball RW, Michell SL. 2014. Lipoprotein CD0873 is a novel adhesin of Clostridium difficile. J Infect Dis 210:274–284. 10.1093/infdis/jiu070. [DOI] [PubMed] [Google Scholar]
- 11.McKee RW, Aleksanyan N, Garrett EM, Tamayo R. 2018. Type IV pili promote Clostridium difficile adherence and persistence in a mouse model of infection. Infect Immun 86:e000943-17. 10.1128/IAI.00943-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piepenbrink KH, Maldarelli GA, de la Pena CF, Mulvey GL, Snyder GA, De Masi L, von Rosenvinge EC, Gunther S, Armstrong GD, Donnenberg MS, Sundberg EJ. 2014. Structure of Clostridium difficile PilJ exhibits unprecedented divergence from known type IV pilins. J Biol Chem 289:4334–4345. 10.1074/jbc.M113.534404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tulli L, Marchi S, Petracca R, Shaw HA, Fairweather NF, Scarselli M, Soriani M, Leuzzi R. 2013. CbpA: a novel surface exposed adhesin of Clostridium difficile targeting human collagen. Cell Microbiol 15:1674–1687. 10.1111/cmi.12139. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Cheng Y, Du P, Zhang Y, Jia H, Li X, Wang J, Han N, Qiang Y, Chen C, Lu J. 2017. Genomic study of the Type IVC secretion system in Clostridium difficile: understanding C. difficile evolution via horizontal gene transfer. Genome 60:8–16. 10.1139/gen-2016-0053. [DOI] [PubMed] [Google Scholar]
- 15.Li N, Jia H, Yang H, Ji B, Liu Y, Peng X, Cheng Y, Zhang W. 2017. Preliminary screening of type IV secretion system in divergent geographic sources of Clostridium difficile. Exp Ther Med 14:4405–4410. 10.3892/etm.2017.5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, Thomson NR, Roberts AP, Cerdeno-Tarraga AM, Wang H, Holden MT, Wright A, Churcher C, Quail MA, Baker S, Bason N, Brooks K, Chillingworth T, Cronin A, Davis P, Dowd L, Fraser A, Feltwell T, Hance Z, Holroyd S, Jagels K, Moule S, Mungall K, Price C, Rabbinowitsch E, Sharp S, Simmonds M, Stevens K, Unwin L, Whithead S, Dupuy B, Dougan G, Barrell B, Parkhill J. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet 38:779–786. 10.1038/ng1830. [DOI] [PubMed] [Google Scholar]
- 17.Brouwer MS, Warburton PJ, Roberts AP, Mullany P, Allan E. 2011. Genetic organisation, mobility and predicted functions of genes on integrated, mobile genetic elements in sequenced strains of Clostridium difficile. PLoS One 6:e23014. 10.1371/journal.pone.0023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grohmann E, Christie PJ, Waksman G, Backert S. 2018. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol 107:455–471. 10.1111/mmi.13896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galan JE, Waksman G. 2018. Protein-injection machines in bacteria. Cell 172:1306–1318. 10.1016/j.cell.2018.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costa TRD, Harb L, Khara P, Zeng L, Hu B, Christie PJ. 2021. Type IV secretion systems: advances in structure, function, and activation. Mol Microbiol 115:436–452. 10.1111/mmi.14670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H, Kubori T, Yamazaki K, Kwak MJ, Park SY, Nagai H, Vogel JP, Oh BH. 2020. Structural basis for effector protein recognition by the Dot/Icm type IVB coupling protein complex. Nat Commun 11:2623. 10.1038/s41467-020-16397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandran Darbari V, Waksman G. 2015. Structural biology of bacterial type IV secretion systems. Annu Rev Biochem 84:603–629. 10.1146/annurev-biochem-062911-102821. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Shen X, Yan J, Han H, Zheng B, Liu D, Cheng H, Zhao Y, Rao X, Wang C, Tang J, Hu F, Gao GF. 2011. GI-type T4SS-mediated horizontal transfer of the 89K pathogenicity island in epidemic Streptococcus suis serotype 2. Mol Microbiol 79:1670–1683. 10.1111/j.1365-2958.2011.07553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Rong C, Chen C, Gao GF. 2012. Type-IVC secretion system: a novel subclass of type IV secretion system (T4SS) common existing in Gram-positive genus Streptococcus. PLoS One 7:e46390. 10.1371/journal.pone.0046390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y, Liu G, Li S, Wang M, Song J, Wang J, Tang J, Li M, Hu F. 2011. Role of a type IV-like secretion system of Streptococcus suis 2 in the development of streptococcal toxic shock syndrome. J Infect Dis 204:274–281. 10.1093/infdis/jir261. [DOI] [PubMed] [Google Scholar]
- 26.Jiang X, Yang Y, Zhou J, Zhu L, Gu Y, Zhang X, Li X, Fang W. 2016. Roles of the putative type IV-like secretion system key component VirD4 and PrsA in pathogenesis of Streptococcus suis type 2. Front Cell Infect Microbiol 6:172. 10.3389/fcimb.2016.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Feng Y, Wang C, Srinivas S, Chen C, Liao H, He E, Jiang S, Tang J. 2017. Pathogenic Streptococcus strains employ novel escape strategy to inhibit bacteriostatic effect mediated by mammalian peptidoglycan recognition protein. Cell Microbiol 19:12724. 10.1111/cmi.12724. [DOI] [PubMed] [Google Scholar]
- 28.Yin S, Li M, Rao X, Yao X, Zhong Q, Wang M, Wang J, Peng Y, Tang J, Hu F, Zhao Y. 2016. Subtilisin-like protease-1 secreted through type IV secretion system contributes to high virulence of Streptococcus suis 2. Sci Rep 6:27369. 10.1038/srep27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallden K, Williams R, Yan J, Lian PW, Wang L, Thalassinos K, Orlova EV, Waksman G. 2012. Structure of the VirB4 ATPase, alone and bound to the core complex of a type IV secretion system. Proc Natl Acad Sci USA 109:11348–11353. 10.1073/pnas.1201428109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabel C, Grahn AM, Lurz R, Lanka E. 2003. The VirB4 family of proposed traffic nucleoside triphosphatases: common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J Bacteriol 185:1045–1058. 10.1128/JB.185.3.1045-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tato I, Zunzunegui S, de la Cruz F, Cabezon E. 2005. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA-dependent ATPase. Proc Natl Acad Sci USA 102:8156–8161. 10.1073/pnas.0503402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Umrekar TR, Cohen E, Drobnic T, Gonzalez-Rodriguez N, Beeby M. 2021. CryoEM of bacterial secretion systems: a primer for microbiologists. Mol Microbiol 115:366–382. 10.1111/mmi.14637. [DOI] [PubMed] [Google Scholar]
- 33.Vermassen A, Talon R, Andant C, Provot C, Desvaux M, Leroy S. 2019. Cell-wall hydrolases as antimicrobials against Staphylococcus species: focus on Sle1. Microorganisms 7:559. 10.3390/microorganisms7110559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Son B, Kong M, Ryu S. 2018. The auxiliary role of the amidase domain in cell wall vinding and exolytic activity of Staphylococcal phage endolysins. Viruses 10:284. 10.3390/v10060284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cohen NR, Ross CA, Jain S, Shapiro RS, Gutierrez A, Belenky P, Li H, Collins JJ. 2016. A role for the bacterial GATC methylome in antibiotic stress survival. Nat Genet 48:581–586. 10.1038/ng.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atack JM, Srikhanta YN, Fox KL, Jurcisek JA, Brockman KL, Clark TA, Boitano M, Power PM, Jen FE, McEwan AG, Grimmond SM, Smith AL, Barenkamp SJ, Korlach J, Bakaletz LO, Jennings MP. 2015. A biphasic epigenetic switch controls immunoevasion, virulence and niche adaptation in non-typeable Haemophilus influenzae. Nat Commun 6:7828. 10.1038/ncomms8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Low DA, Weyand NJ, Mahan MJ. 2001. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun 69:7197–7204. 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliveira PH, Fang G. 2021. Conserved DNA methyltransferases: a window into fundamental mechanisms of epigenetic regulation in bacteria. Trends Microbiol 29:28–40. 10.1016/j.tim.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oliveira PH, Ribis JW, Garrett EM, Trzilova D, Kim A, Sekulovic O, Mead EA, Pak T, Zhu S, Deikus G, Touchon M, Lewis-Sandari M, Beckford C, Zeitouni NE, Altman DR, Webster E, Oussenko I, Bunyavanich S, Aggarwal AK, Bashir A, Patel G, Wallach F, Hamula C, Huprikar S, Schadt EE, Sebra R, van Bakel H, Kasarskis A, Tamayo R, Shen A, Fang G. 2020. Epigenomic characterization of Clostridioides difficile finds a conserved DNA methyltransferase that mediates sporulation and pathogenesis. Nat Microbiol 5:166–180. 10.1038/s41564-019-0613-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arechaga I, Pena A, Zunzunegui S, del Carmen Fernandez-Alonso M, Rivas G, de la Cruz F. 2008. ATPase activity and oligomeric state of TrwK, the VirB4 homologue of the plasmid R388 type IV secretion system. J Bacteriol 190:5472–5479. 10.1128/JB.00321-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tato I, Matilla I, Arechaga I, Zunzunegui S, de la Cruz F, Cabezon E. 2007. The ATPase activity of the DNA transporter TrwB is modulated by protein TrwA: implications for a common assembly mechanism of DNA translocating motors. J Biol Chem 282:25569–25576. 10.1074/jbc.M703464200. [DOI] [PubMed] [Google Scholar]
- 42.Durand E, Oomen C, Waksman G. 2010. Biochemical dissection of the ATPase TraB, the VirB4 homologue of the Escherichia coli pKM101 conjugation machinery. J Bacteriol 192:2315–2323. 10.1128/JB.01384-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hormaeche I, Alkorta I, Moro F, Valpuesta JM, Goni FM, De La Cruz F. 2002. Purification and properties of TrwB, a hexameric, ATP-binding integral membrane protein essential for R388 plasmid conjugation. J Biol Chem 277:46456–46462. 10.1074/jbc.M207250200. [DOI] [PubMed] [Google Scholar]
- 44.Rangrez AY, Abajy MY, Keller W, Shouche Y, Grohmann E. 2010. Biochemical characterization of three putative ATPases from a new type IV secretion system of Aeromonas veronii plasmid pAC3249A. BMC Biochem 11:10. 10.1186/1471-2091-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pena A, Matilla I, Martin-Benito J, Valpuesta JM, Carrascosa JL, de la Cruz F, Cabezon E, Arechaga I. 2012. The hexameric structure of a conjugative VirB4 protein ATPase provides new insights for a functional and phylogenetic relationship with DNA translocases. J Biol Chem 287:39925–39932. 10.1074/jbc.M112.413849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker JE, Saraste M, Runswick MJ, Gay NJ. 1982. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1:945–951. 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomis-Rüth FX, Moncalián G, Pérez-Luque R, González A, Cabezón E, de la Cruz F, Coll M. 2001. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature 409:637–641. 10.1038/35054586. [DOI] [PubMed] [Google Scholar]
- 48.Moncalian G, Cabezon E, Alkorta I, Valle M, Moro F, Valpuesta JM, Goni FM, de La Cruz F. 1999. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J Biol Chem 274:36117–36124. 10.1074/jbc.274.51.36117. [DOI] [PubMed] [Google Scholar]
- 49.Panicker MM, Minkley EG. 1992. Purification and properties of the F sex factor TraD protein, an inner membrane conjugal transfer protein. J Biol Chem 267:12761–12766. 10.1016/S0021-9258(18)42341-1. [DOI] [PubMed] [Google Scholar]
- 50.Schroder G, Krause S, Zechner EL, Traxler B, Yeo HJ, Lurz R, Waksman G, Lanka E. 2002. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol 184:2767–2779. 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schroder G, Lanka E. 2003. TraG-like proteins of type IV secretion systems: functional dissection of the multiple activities of TraG (RP4) and TrwB (R388). J Bacteriol 185:4371–4381. 10.1128/JB.185.15.4371-4381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J, Lee HJ, Shin MK, Ryu WS. 2004. Versatile PCR-mediated insertion or deletion mutagenesis. Biotechniques 36:398–400. 10.2144/04363BM04. [DOI] [PubMed] [Google Scholar]
- 53.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gribling-Burrer AS, Chiabudini M, Zhang Y, Qiu Z, Scazzari M, Wolfle T, Wohlwend D, Rospert S. 2019. A dual role of the ribosome-bound chaperones RAC/Ssb in maintaining the fidelity of translation termination. Nucleic Acids Res 47:7018–7034. 10.1093/nar/gkz334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 56.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA 76:4350–4354. 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nesterenko MV, Tilley M, Upton SJ. 1994. A simple modification of Blum's silver stain method allows for 30 minute detection of proteins in polyacrylamide gels. J Biochem Biophys Methods 28:239–242. 10.1016/0165-022X(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 58.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S9 and Tables S1 and S2. Download JB.00359-21-s0001.pdf, PDF file, 1.0 MB (1,019.1KB, pdf)