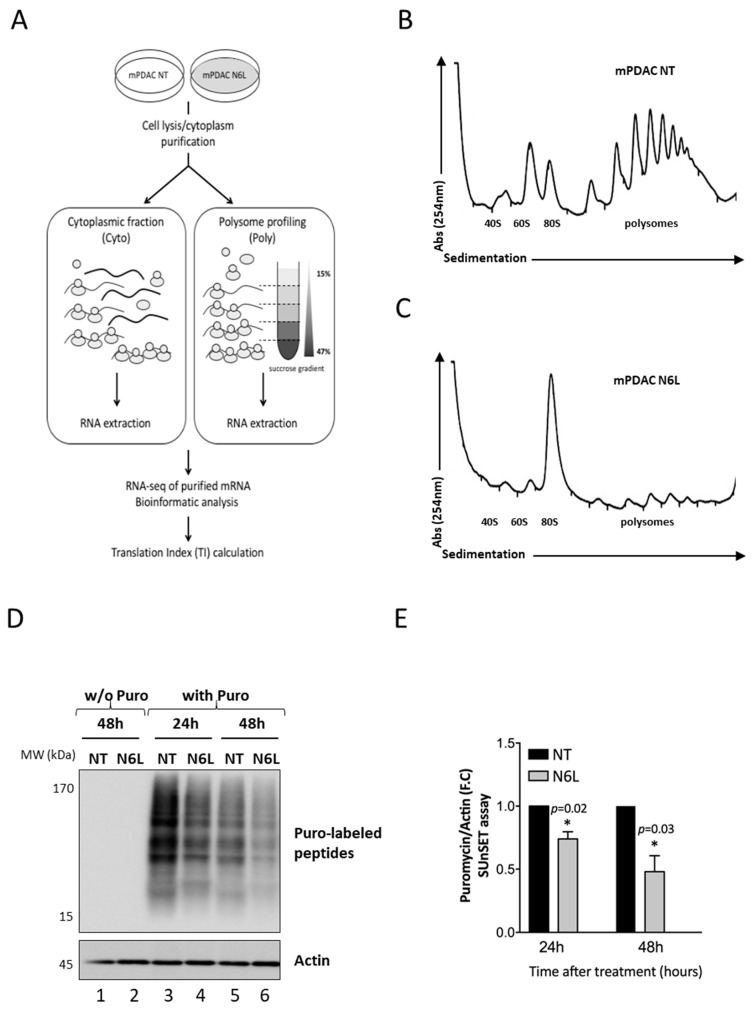
Figure 1.
NCL targeting by N6L impacts mPDAC cell translation. (A)—Experimental design of translatome analysis procedure. mPDAC cells were treated (T) or not (NT) with 50 µM of N6L for 48 h. Cytosolic lysates from cells were sedimented on a sucrose gradient (15–47%). Following ultracentrifugation, 40S and 60S ribosome subunits, the 80S monosomes, and polysomes are separated. RNAs were extracted from polysome pooled fractions (mRNA-associated polysomes) and cytoplasmic fractions (cytoplasmic mRNA). The RNA quality and integrity were verified using a Bioanalyser 2100 before sequencing and bioinformatics analysis. (B,C)—Polysome profiles of mPDAC cells non-treated (mPDAC NT) or treated (mPDAC T) with N6L. Curves show absorbance at 254 nm as a function of sedimentation. (D,E)—mPDAC cells, treated or not with N6L, were incubated with puromycin for the measurement of overall protein synthesis by the SUnSET method and then subjected to Western blotting (D). Protein synthesis rates were quantified by measuring the signal intensity in each lane and normalizing the values to that of the control lane (E). w/o Puro, without puromycin. Data represented are the means ± SD of at least three experiments. * p < 0.05 and n.s. for not significant, as determined by Student’s t-test on final data point.