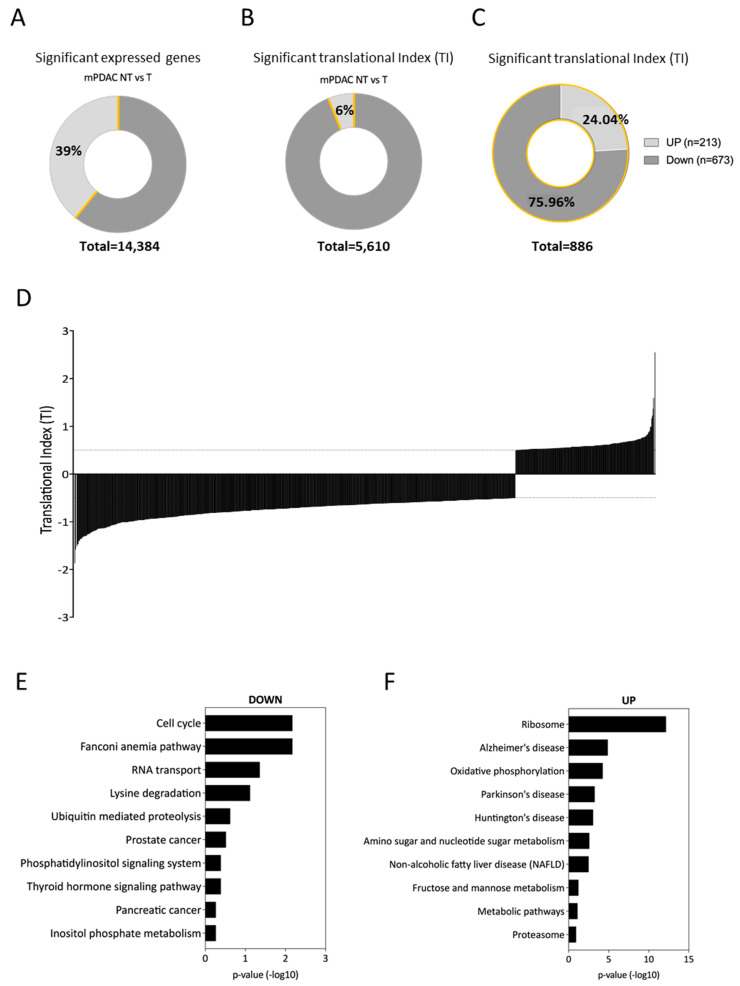
Figure 2.
Bioinformatics analysis of mPDAC cell translatome upon N6L treatment. A—Percentage of translationally deregulated mRNAs in response to N6L treatment. Among the 14,384 blasted genes, 39% (5610) were significantly expressed in both NT and T conditions (A). About 6% (886) of these 5610 genes were significantly deregulated at translational levels (TI cut-off = 1.5, p < 0.05) (B). Among these 886 translationally deregulated genes, 24.04% were up-regulated, and 75.96% were down-regulated (C). Distribution of the Translational Index (TI) (TI = X1/X2), with X1 and X2 expressing the ratio between polysomal-to-cytoplasmic fraction in mPDAC-treated and non-treated cells, respectively (D) (complete gene list in Table S1). (E,F)—Ontology analysis was determined using the functional annotation clustering tools from EnrichR software. Translationally down-regulated genes are involved in the cell cycle, Fanconi anemia and RNA transport (E). While translationally up-regulated genes are mainly involved in ribosome/translation and metabolism (F).