Figure 2.
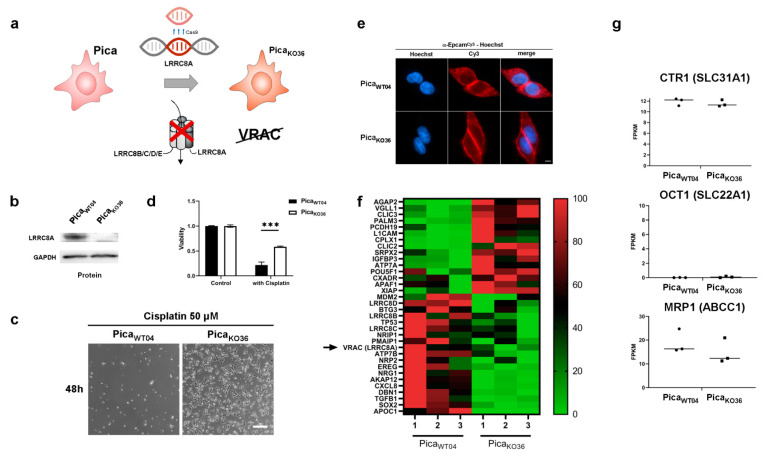
Profiling cisplatin sensitivity pathways underlines the relevance of VRAC as critical for cisplatin resistance. (a) Illustration of CRISPR/Cas9 technology to establish the VRAC-deficient, cisplatin-resistant, knockout cell line (PicaKO36). Scheme of the VRAC channel, consisting of six heteromeric subunits, indicated (b) Western blot to confirm the absence of LRRC8A protein expression in PicaKO36 cells. GAPDH served as loading control. (c) In contrast to WT cells, the VRAC-deficient cell line PicaKO36 is able to survive treatment with high cisplatin concentrations. Cells were treated with cisplatin (50 µM) for 48 h and imaged by live cell microscopy. Scale bar, 200 µm. (d) In contrast to WT cells, the VRAC-deficient cell line, PicaKO36, is significantly cisplatin resistant. Cells were exposed to cisplatin (20 µM) for 48 h and viability was normalized to untreated controls. ***, p < 0.005 (e) PicaWT04 and PicaKO36 cells show similar morphology. Fluorescence microscopy visualized EpCAM expression (stained with specific antibodies (red)), nuclei were stained with Hoechst (blue). Scale bar, 5 µm. (f) Applying RNASeq transcriptomics to identify cisplatin resistance players. Heatmap visualizing expression levels of potential cisplatin resistance-associated genes, which are differentially expressed in KO (PicaKO36) vs. cisplatin sensitive WT (PicaWT04) cells (green: downregulated, red: upregulated; full list of differentially expressed genes in Table S4). Constituting subunit of VRAC (LRRC8A) indicated. (g) Expression levels of previously described cisplatin-resistance channel proteins (CTR1, OCT1, MRP1; see Figure 1b) are unaffected in the HNSCC knockout cell line and thus are less relevant for HNSCC. RNA intensities as FPKM values are displayed. For uncropped blots, refer to Supplementary Materials.