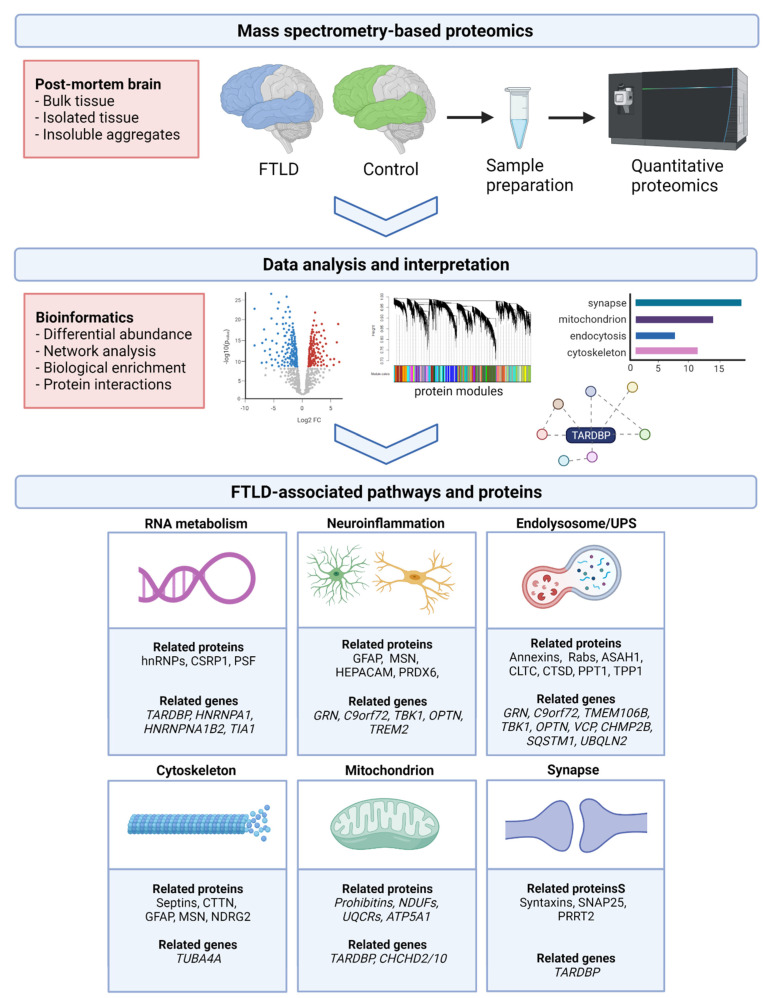
Figure 1.
Unbiased proteomics to characterize the proteomic signature of FTLD-TDP. Following brain tissue preparation, several techniques are currently available to quantify the proteomic data generated by mass spectrometry. Different bioinformatics approaches can be applied to analyze the differentially expressed proteome; i.e., to organize these proteins in biologically meaningful groups (modules), to evaluate the protein-protein interactions, or to assess enrichment for specific biological pathways, cell types, and/or genetic risk factors. By reviewing the various proteomic studies performed in FTLD-TDP, six core pathways could be identified that are implicated in the disease pathogenesis. For each pathway, we mention several central proteins/protein families, which were either detected by multiple studies or validated by individual studies (see also Table 2 and Table S1). In addition, the major FTLD genes associated with these pathways are listed, as derived from literature. Importantly, both lists should be regarded as examples and are not exhaustive. Created with BioRender.com.