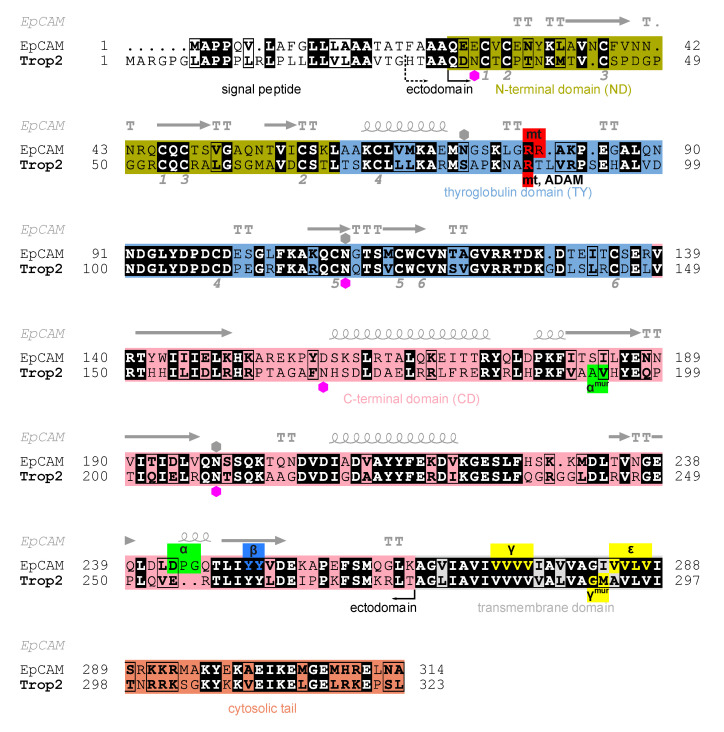
Figure 1.
Alignment of human Trop2 (UniProt P09758-1) and EpCAM (UniProt P16422-1) amino acid sequences with marked N-terminal (olive), thyroglobulin (light blue), C-terminal (pink), transmembrane (grey) and cytosolic (orange) regions/domains. Identical residues are shown in bold and with black background, and chemically similar residues in bold and in rectangles. Hexagons denote N-glycosylation sites as annotated in UniProt for Trop2 (magenta) and EpCAM (grey). Pairs of cysteine residues forming a disulfide bridge are denoted by equal numbers below the alignment (from one to six, corresponding to six disulfide bridges). Secondary structure elements in EpCAM (PDB ID 4MZV [6]) are shown above EpCAM sequence: α-helices (spiral), β-strands (arrows), α-turns (TTT) and β-turns (TT). Smaller red, green, blue and yellow boxes denote experimentally determined proteolytic cleavage sites on human Trop2 and EpCAM, by matriptase (mt), TACE (α), ADAM10 (ADAM), β-secretase 1 (BACE, β) and γ-secretase (γ, ε). Cleavage sites determined using a murine protein are denoted by “mur” superscript; the corresponding residues in human protein were deduced from alignment of murine and human sequences. Two alternative N-termini of the ectodomain are marked: H27 (currently annotated in UniProt) and Q31 (predicted using SignalP [25]). Figure was prepared using ESPript 3.0 [26], and the output has been manually edited to include features and topological regions.