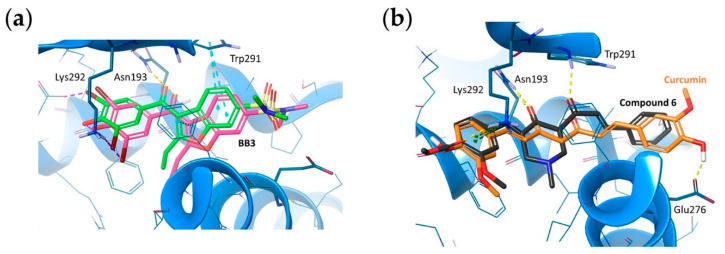
Figure 8.
(a) Spatial orientation of BB3 within the allosteric site of PtP1B in the original crystal structure (PDB ID: 1T48; green ligand) and docked to the obtained, optimized model (pink ligand). RMSD obtained for BB3 redocking: 1.66 Å. CBR1 backbone displayed as blue ribbon. Amino acid residues engaged in ligand binding throughout H-bonds (dotted yellow lines), π-interactions (dotted blue lines), and halogen bonds (dotted purple lines) are displayed as sticks. (b) The predicted binding mode comparison of compound 6 (black) and curcumin (orange) within the allosteric site of PTP1B. Amino acid residues within 5 Å displayed as thin, blue sticks; amino acid residues engaged in ligand binding throughout H-bonds (dotted yellow lines) and π-cation interaction (dotted green lines) displayed as bold, blue sticks.