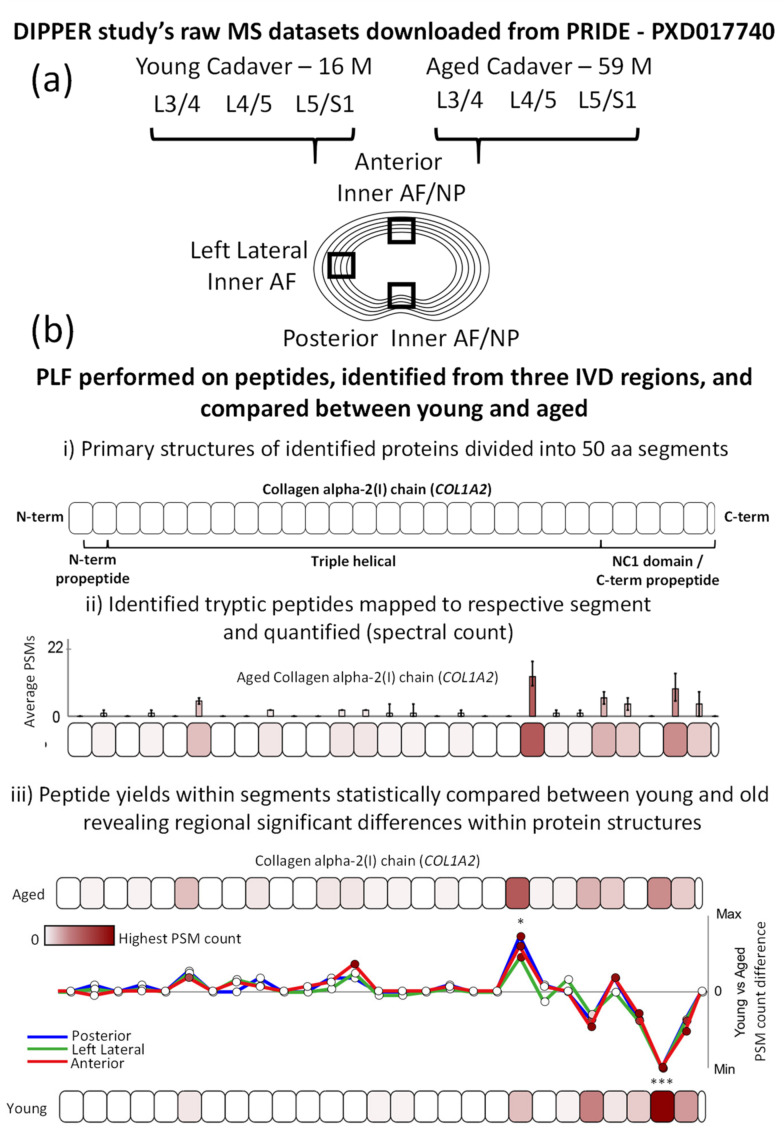
Figure 1.
Experimental design for the application of PLF to three distinct regions of young and aged IVDs. (a) Raw LC–MS/MS datasets (from two males, one young [16 years old] and one aged [59 years old]), corresponding to the posterior inner AF/NP, left lateral inner AF and anterior inner AF/NP regions of the L3/4, L4/5 and L5/S1 lower lumbar IVDs were downloaded from the PRIDE repository (PXD017740) [13]. (b) After MS/MS peptide identification, the MPLF webtool [25] was used to perform PLF on the identified peptides and compare peptide yields across protein structures between young and aged discs—three vs. three discs for anterior and left lateral datasets and three vs. two discs for posterior datasets (the young anterior L4/5 disc was omitted from the analysis due to the insufficient number of identified peptides for PLF analysis). As previously described [25], ((b), i) proteins were divided into 50-amino acid (aa)-sized segments; the collagen I α2 chain (COL1A2) is shown here as an example. ((b), ii) Peptides were mapped to their respective segments, summed, normalised between aged and young groups based on the median whole protein spectrum count and averaged for each group (average peptide spectrum match [PSM] count per segment). ((b), iii) Peptide yields in each segment were statistically compared between young and aged IVDs using an unpaired, Bonferroni-corrected, repeated-measures ANOVA (* p ≤ 0.05; *** p ≤ 0.001). To visualise these differences along the protein structure, average PSM counts per segment in the young group were subtracted from those in the aged one and compared between posterior, left lateral and anterior IVD regions.