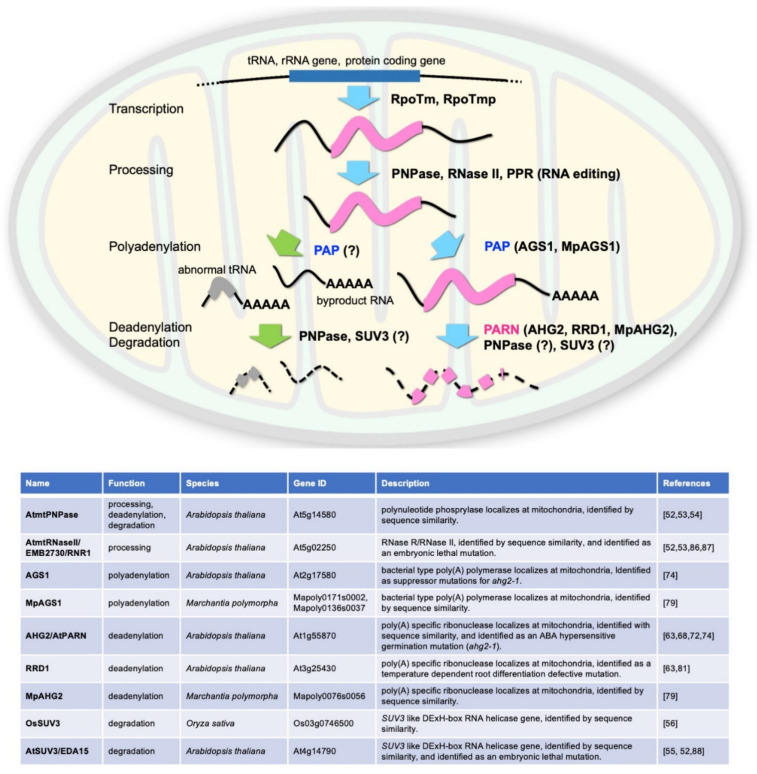
Figure 1.
Processing of plant mitochondrial mRNAs. Upper panel: In land plant mitochondria, genes in the mitochondrial genome are transcribed separately by T3/T7 phage type RNA polymerases (RpoTm and RpoTmp) [26,27,28]. Primary transcripts of protein coding mRNAs are trimmed at their 5′ and 3′ ends by polynucleotide phosphorylase (PNPase) and RNase II [26,27,29,30]. Some mRNA is edited at specific residues by pentatricopeptide repeat proteins (PPRs). Unknown regulatory signals then induce polyadenylation at the 3′ ends of the mRNAs via a poly(A) polymerase (PAP) such as AHG2 SUPPRESSOR1 (AGS1) [31]. RNAs with abnormal structures are degraded by the mRNA quality surveillance machinery. Polyadenylated mRNA is believed to be degraded by RNase following its deadenylation by poly(A)-specific ribonucleases (PARNs) such as ABA-HYPERSENSITIVE GERMINATION2 (AHG2)/AtPARN [31,32,33,34] and ROOT REDIFFERENTIATION DEFECTIVE1 (RRD1) [32,35]. PNPase may participate in this process. MpAGS1 and MpAHG2 are Marchantia paralogs of Arabidopsis AGS1 and AHG2 [36], respectively. SUV3-like proteins may be involved in certain processes (right side) [26,37,38,39]. tRNAs (and rRNAs) with abnormal structures and byproducts of mRNA processing are also polyadenylated and degraded, presumably by PNPase and the SUV3-like protein complex, as observed in human mitochondria (left side). Lower panel: the information of the factors shown in the upper panel. ? mark means that the involvement of these factors have been suggested but not confirmed yet.