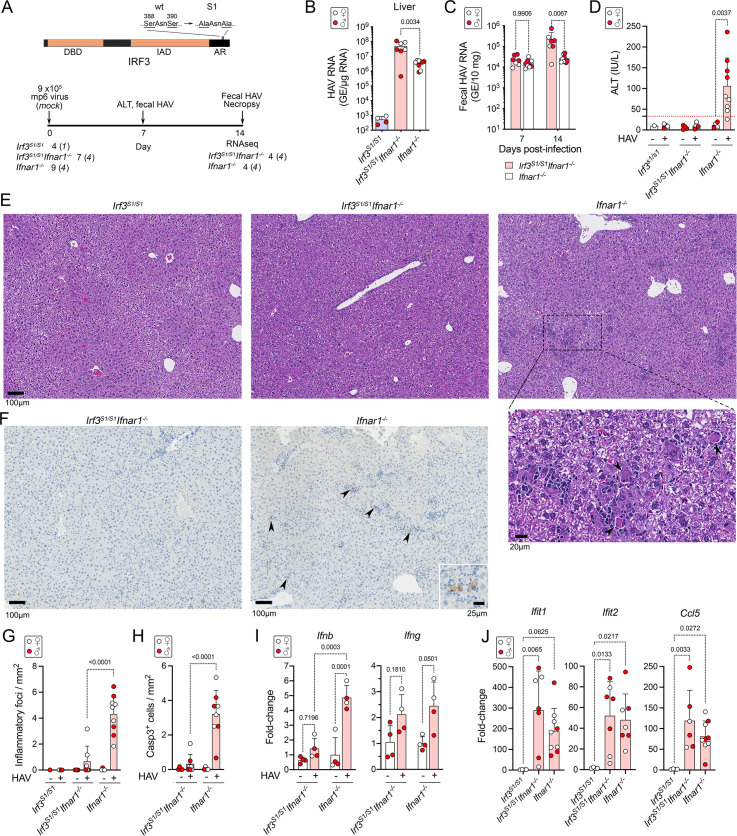
Fig 3. Infectious HAV challenge of Irf3S1/S1, Irf3S1/S1Ifnar1-/-, and Ifnar1-/- mice.
(A) (top) IRF3 protein structure showing location of Ser-to-Ala substitutions in Irf3S1/S1 mice that render the protein transcriptionally inactive. DBD, DNA-binding domain; IAD, IRF-association domain; AR, autoinhibitory region. (bottom) Experimental scheme. A total of 4 Irf3SI/SI mice, 7 Irf3S1/S1Ifnar1-/- mice, and 9 Ifnar1-/- mice were infected with 9 x105 HM175-mp6 virus in two independent experiments. Liver tissue collected at necropsy 14 days later from representative infected and mock-infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice (4 each) were selected for high-throughput RNA sequencing. Irf3SI/SI mice showed no evidence of infection and these livers were not subjected to sequencing. (B) HAV RNA in livers of mice determined by RT-qPCR 14 days post-inoculation (dpi). In all panels, male animals are indicated by solid red symbols, and female animals by open symbols. (C) Fecal HAV RNA 7 and 14 dpi. (D) Serum ALT in Irf3S1/S1, Irf3S1/S1Ifnar1-/-, and Ifnar1-/- mice 7 dpi. (E) Representative H&E-stained sections of (left to right) Irf3S1/S1, Irf3S1/S1Ifnar1-/-, and Ifnar1-/- mouse liver 14 dpi. Arrowheads in the enlarged image of Ifnar1-/- liver indicate apoptotic hepatocytes. (F) Representative immunohistochemically stained liver sections showing detection of cleaved caspase 3 in liver from (left) Irf3S1/S1Ifnar1-/- and (right) Ifnar1-/- mice 14 dpi. (G) Number of inflammatory foci per mm2 in H&E-stained liver sections. (H) Mean number of hepatocytes staining positively for cleaved caspase 3 (Casp3) per mm2 in liver sections by immunohistochemistry. (I) RT-qPCR determination of the fold-change in intrahepatic Ifnb and Ifng transcript abundance in groups of infected and mock-infected Irf3S1/S1Ifnar1-/- and Ifnar1-/- mice selected for high-throughput sequencing (n = 4 in each group). (J) RT-qPCR determination of infection-induced changes in intrahepatic Ifit1, Ifit2 and Ccl5 transcript levels in mice from both experiments. Error bars in all panels indicate s.d.; p-values are shown for 2-way comparisons by two-way ANOVA with Šidák’s test for multiple comparisons.