Figure 3.
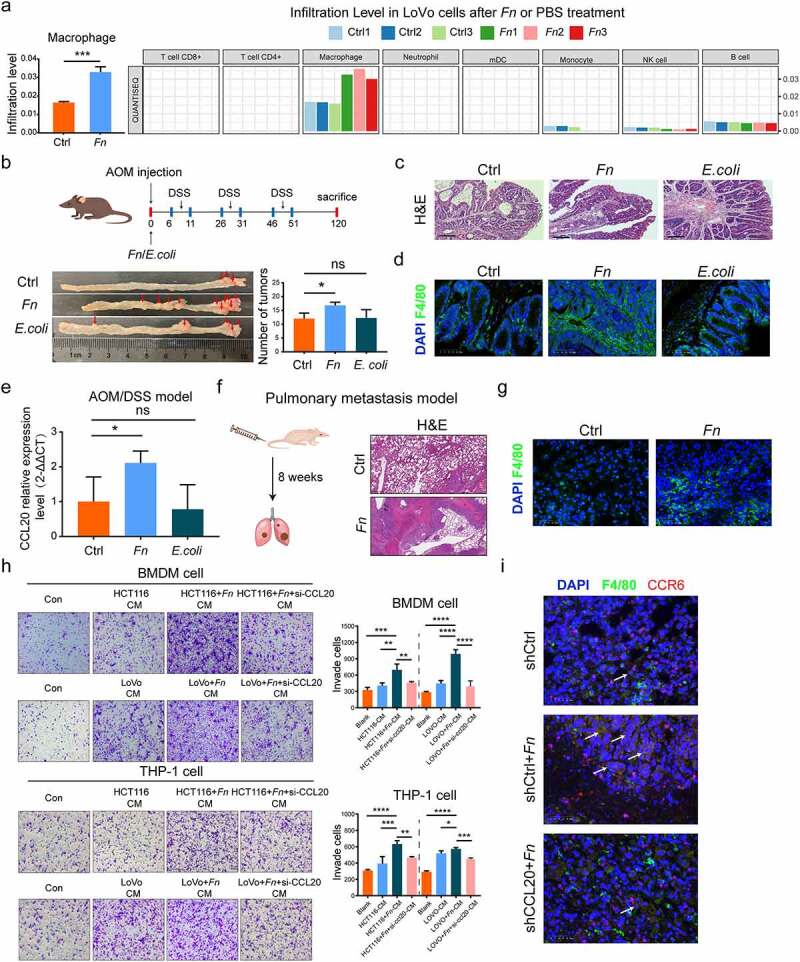
Tumor-derived CCL20 induced by F. nucleatum mediated the macrophage infiltration in the TME. (a) The immune cells infiltration was analyzed via TIMER 2.0 and quanTIseq based on RNA-seq profiles. (b) Colon tumor number in AOM/DSS model was counted in F. nucleatum or PBS or E. coli groups (Student’s test; n = 4–8 per group). The red arrows indicated the tumor locations. (c) Representative images of H&E staining in the intestinal tissues in AOM/DSS CRC model after F. nucleatum or PBS or E. coli treatment. Scale bars:500 μm. (d) The proportion of F4/80+ macrophage was detected by immunofluorescence in AOM/DSS CRC model after F. nucleatum or PBS or E. coli treatment (Student’s t test). Scale bars:25 μm. (e) The CCL20 expression was detected in AOM/DSS model after F. nucleatum or PBS or E. coli treatment (Student’s t test). (f) Representative images of H&E staining in the pulmonary metastasis model after F. nucleatum or PBS treatment. Scale bars:200 μm. (g)The proportion of F4/80+ macrophage was detected by immunofluorescence in the pulmonary metastasis model after F. nucleatum or PBS treatment. Scale bars:25 μm. (h) After co-cultured with culture medium of CRC cells pretreated by F. nucleatum and/or si-RNA CCL20 for 24 h, the mobility of macrophage was detected by transwell assay (Each experiment was repeated in triplicate; Student’s t test). Scale bars:100 μm. (i) Representative images of the F4/80+ CCR6+ macrophage in the pulmonary metastasis model after control shRNA or control shRNA+ F. nucleatum or CCL20 shRNA+ F. nucleatum treatment were detected by immunohistochemistry. The white arrows indicated F4/80+ CCR6+ macrophage. Scale bars:25 μm. * p < .05, ** p < .01, and *** p < .001, **** p < .0001, ns no significant
