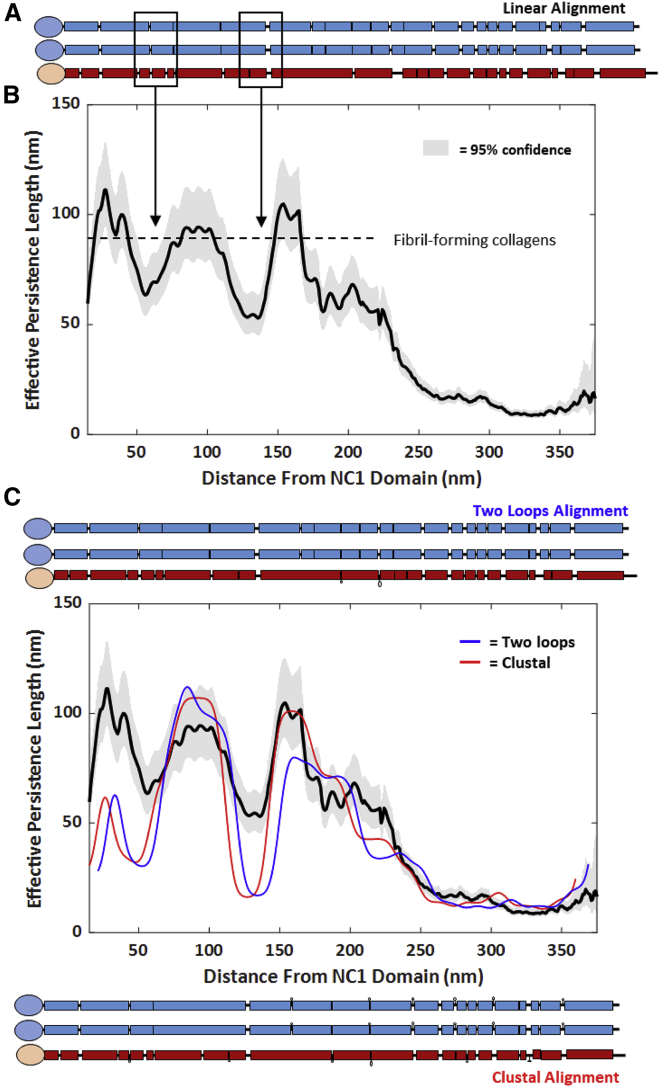
Figure 4.
Position-dependent flexibility profile of collagen IV. (A) Schematic representation of the α1 and α2 amino acid sequences from mouse collagen type IV. Rectangles indicate regions of the sequence containing triple-helix-competent sequences with (Gly-X-Y)nG, n ≥ 4. Interruptions of this repetitive sequence are indicated by thinner lines and occur more frequently toward the N-terminus of the chains (right side of the schematic). (B) Position-dependent persistence length map of collagen type IV deposited from 100 mM KCl, 1 mM HCl. The dashed line indicates the persistence length of continuously triple-helical fibril-forming collagen molecules (10). Shaded curves represent 95% confidence intervals on the effective persistence length estimate p∗(s; Δs). The profile was calculated from n = 262 chains. The effective persistence length map begins ~15 nm into the collagen sequence because of the use of centered 30-nm windows for determination of p∗. (C) Four-class flexibility model using two-loop (blue) and Clustal (red) alignments. Alignments differ in the number of amino acids that loop out and thus do not participate in the main backbone, as shown schematically above and below the plot. The persistence length profile is aligned with the amino acid sequence representations using model outputs for best-fit offset (nm) and nanometer/amino acid conversion. The two-loop (Clustal) alignment is best fitted with persistence lengths of p0 = 105 (109) nm, p1 = 83 (86) nm, p2 = 21 (42) nm, and p3 = 1.9 (2.0) nm, with χ2r = 12.6 (12.8). Best-fit persistence lengths of these different structural classes, particularly p1 and p2, differ for other chain alignments (Table S1). To see this figure in color, go online.