Fig. 4. H3K27me3 is dynamically changed during mESC division with Wnt3a beads.
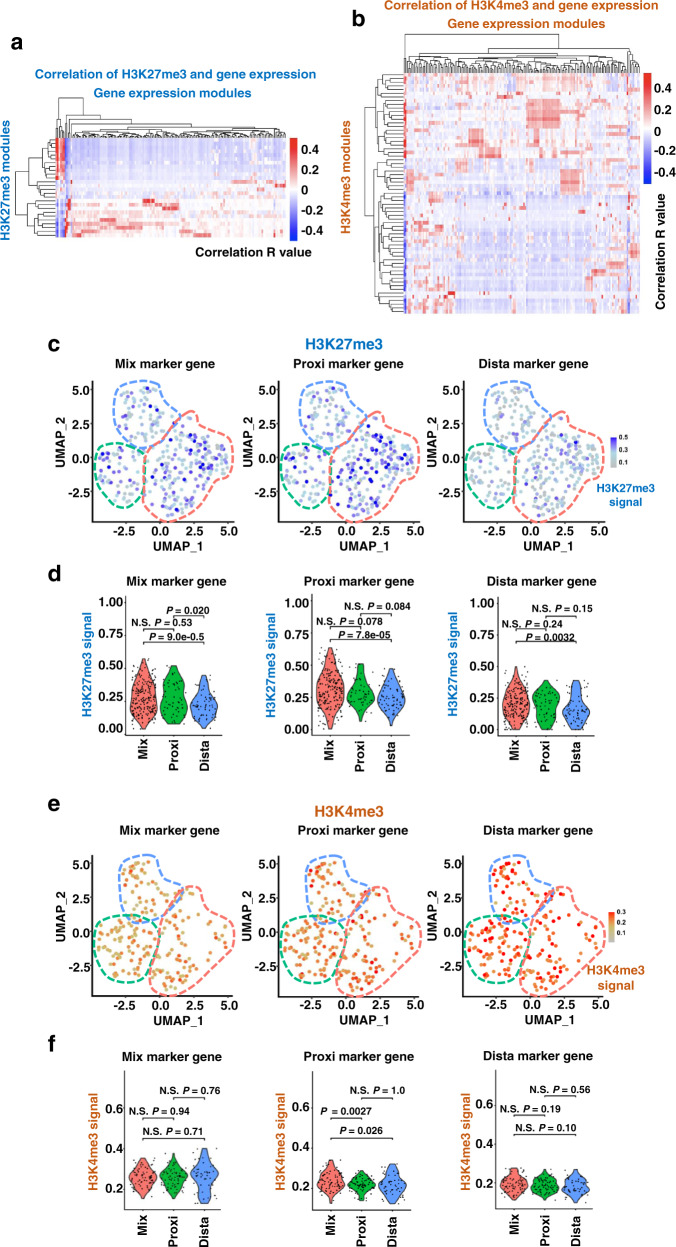
a Heatmap showing the correlations between H3K27me3 modules and transcriptional modules defined by WGCNA. Pearson correlation coefficient was calculated by module scores. b Heatmap showing the correlations between H3K4me3 modules and transcriptional modules defined by WGCNA. Pearson correlation coefficient was calculated by module scores. c Enrichments of H3K27me3 at cluster marker genes in individual cells of H3K27me3 scSET-seq. Total levels of H3K27me3 at the Mix, Proxi, and Dista cluster marker genes were shown respectively. d Violin plots showing the summarized signals in each cluster of cells. Cells were divided into Mix, Proxi, and Dista clusters and shown as the x-axis. H3K27me3 signals at all marker genes from Mix, Proxi, and Dista clusters were shown respectively. P values were calculated by Student’s t-test, two-sided. N.S. Not Significant. n = 208 (H3K27me3 Mix cluster), 58 (H3K27me3 Proxi cluster), and 60 (H3K27me3 Dista cluster) cells. e Same as in c, except H3K4me3 scSET-seq was shown. f Same as in d, except H3K4me3 scSET-seq was shown. P values were calculated by Student’s t-test, two-sided. N.S. Not Significant. n = 83 (H3K4me3 Mix cluster), 75 (H3K4me3 Proxi cluster), and 52 (H3K4me3 Dista cluster) cells. Source data are provided as a Source Data file.