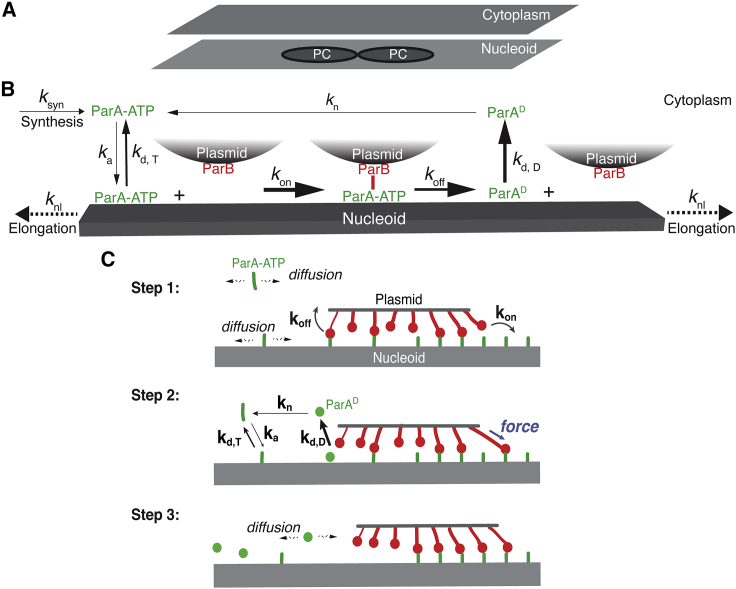
Figure 1.
Model description. (A) Model setup. The model describes each PC as a circular disk and the nucleoid and the cytoplasm as the rectangular domains that share the same dimensions. (B) Biochemical scheme of ParABS system. Briefly, ParB molecules are fixed on the PCs, whereas the ParA·ATP molecules exchange between the cytoplasm and nucleoid surface with the kinetic rates of ka and kd, T, respectively. The PC-bound ParB will rapidly bind to the local nucleoid-bound ParA·ATP at the rate of kon, followed by a fast dissociation at the rate of koff. The dissociation of ParA-ParB bond drives the nucleoid-bound ParA into a distinctive state, ParAD, which rapidly turns over into the cytoplasm at the rate of kd, D. At a very slow rate (kn), the cytoplasmic ParAD will revert into the ParA·ATP that can rebind to the nucleoid. Last, to capture the essential effects of cell growth on the ParABS system, the nucleoid keeps elongating at the rate of knl, and ParA·ATP molecules are synthesized at the rate of ksyn. (C) Mechanochemical coupling of ParA-ParB bond dynamics underlies ParA gradient-based Brownian ratcheting. The Brownian ratcheting consists of three stochastic steps. Step 1: although some of the ParA-ParB bonds are dissociating, others are forming. The free ParA molecules diffuse along the nucleoid and in the cytoplasm, whereas all the ParB molecules are fixed the PC. Step 2: thermal fluctuation prestretches the forming ParA-ParB bond that pulls the PC forward. At the back of the PC, the ParA-ParB bond dissociation converts the ParA·ATP into a distinctive state (ParAD) with a rapid turnover into the cytoplasm. Because of the slow ParAD-to-ParA·ATP conversion, there is a time delay to replenish the local ParA·ATP on the nucleoid. This results in a ParA depletion trailing behind the PC. Step 3: the initial movement of PC creates an asymmetric local ParA concentration gradient (higher at the front, lower at the back). This breaks symmetry and drives the directed and persistent PC movement.