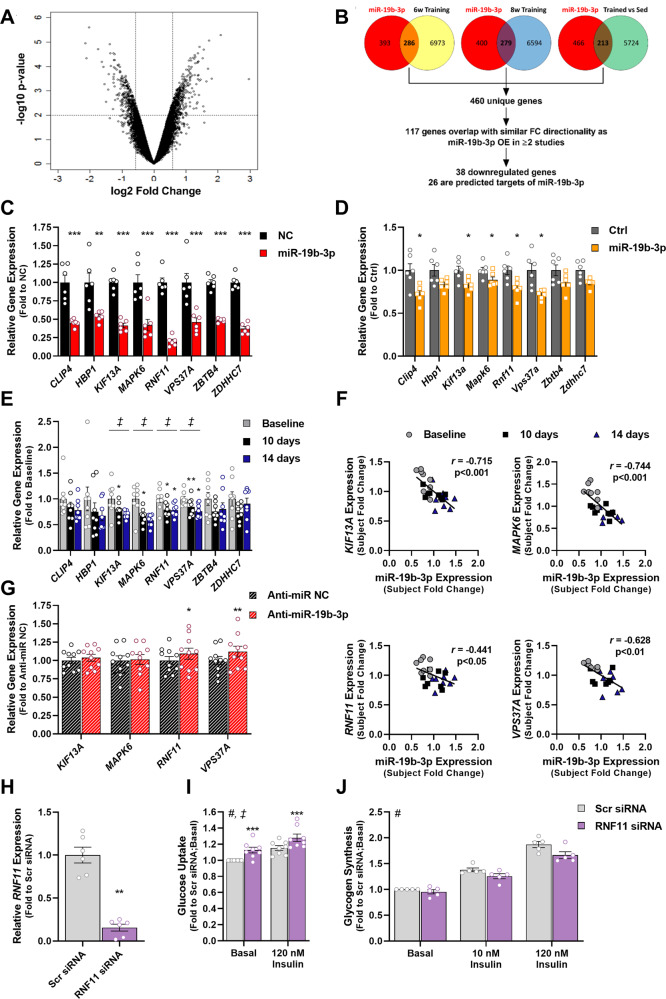
Fig. 4. miR-19b-3p modulates expression of genes altered by endurance exercise training in human skeletal muscle.
A Volcano plot showing changes in gene expression determined by microarray analysis of myotubes overexpressing miR-19b-3p. Dashed lines indicate value cut-off at p < 0.01 and fold change of at least ±50% (n = 4 independent cell culture experiments). B Overview of workflow to identify genes with altered expression profiles following miR-19b-3p overexpression in human skeletal muscle cells and following aerobic exercise training (analysis of publicly available data8,15,16). C Expression of miR-19b-3p predicted target genes CLIP4 (p = 0.0001), HBP1 (p = 0.0032), KIF13A (p = 0.0001), MAPK6 (p = 0.0007), RNF11 (p = 0.0001), VPS37A (p = 0.0004), ZBTB4 (p = 0.0001), and ZDHHC7 (p = 0.0001) as determined by RT-qPCR following negative control (NC) or miR-19b-3p overexpression in human skeletal muscle cells (n = 6 independent cell culture experiments). D Effect of miR-19b-3p overexpression in mouse tibialis anterior skeletal muscle on expression of Clip4 (p = 0.0275), Hbp1, Kif13a (p = 0.0388), Mapk6 (p = 0.0116), Rnf11 (p = 0.034), Vps37a (p = 0.0206), Zbtb4, and Zdhhc7 as determined by RT-qPCR (n = 6 independent mouse samples). E Gene expression of CLIP4, HBP1, KIF13A (p = 0.0497 at day 10), MAPK6 (p = 0.0143 at day 10 and p = 0.0031 at day 14), RNF11 (p = 0.038 at day 10 and p = 0.017 at day 14), VPS37A (p = 0.0051 at day 10 and p = 0.0222 at day 14), ZBTB4, and ZDHHC7 as determined by RT-qPCR in human skeletal muscle biopsies collected before (Baseline) and after 10 or 14 days of endurance exercise training (n = 8 individuals). F Pearson correlation of fold changes in miR-19b-3p expression and fold change of KIF13A (p = 0.0001), MAPK6 (p = 0.0001), RNF11 (p = 0.0312), or VPS37A (p = 0.001) expression in human skeletal muscle biopsies collected prior to (Baseline) and after 10 or 14 days of endurance exercise training (n = 24 biopsies). G Human myotubes were transfected with a miR-19b-3p-specific inhibitor (Anti-miR-19b-3p) or a negative control inhibitor (Anti-miR NC) and gene expression of KIF13A, MAPK6, RNF11 (p = 0.024), and VPS37A (p = 0.0034) was determined by RT-qPCR (n = 10 independent cell culture experiments). H Human skeletal muscle myotubes were transfected with a scramble siRNA sequence (Scr siRNA) or a RNF11 siRNA and gene expression of RNF11 was determined by RT-qPCR (p = 0018, n = 6 independent cell culture experiments). I Human skeletal muscle myotubes were incubated in the absence (Basal, p = 0.0004) or presence of insulin (120 nM, p = 0.0008) to determine the effect of RNF11 silencing on glucose uptake (n = 8 independent cell culture experiments). J Glycogen synthesis rate was determined in human skeletal muscle cells in the absence (Basal) or presence of insulin (10 or 120 nM) following RNF11 silencing (n = 5 independent cell culture experiments). Data are mean ± SEM. RT-qPCR data in human skeletal muscle biopsies and cells were normalized to the geometrical mean of GUSB and TBP expression. For mouse TA muscle, gene expression was normalized to Tbp expression. # Insulin effect; ‡ Endurance exercise training effect; and *p < 0.05, **p < 0.01, ***p < 0.001 by two-tailed paired Student’s t-test, repeated measures one-way ANOVA with Dunnett’s post hoc testing, or repeated measures two-way ANOVA with Sidak’s post hoc testing when appropriate.