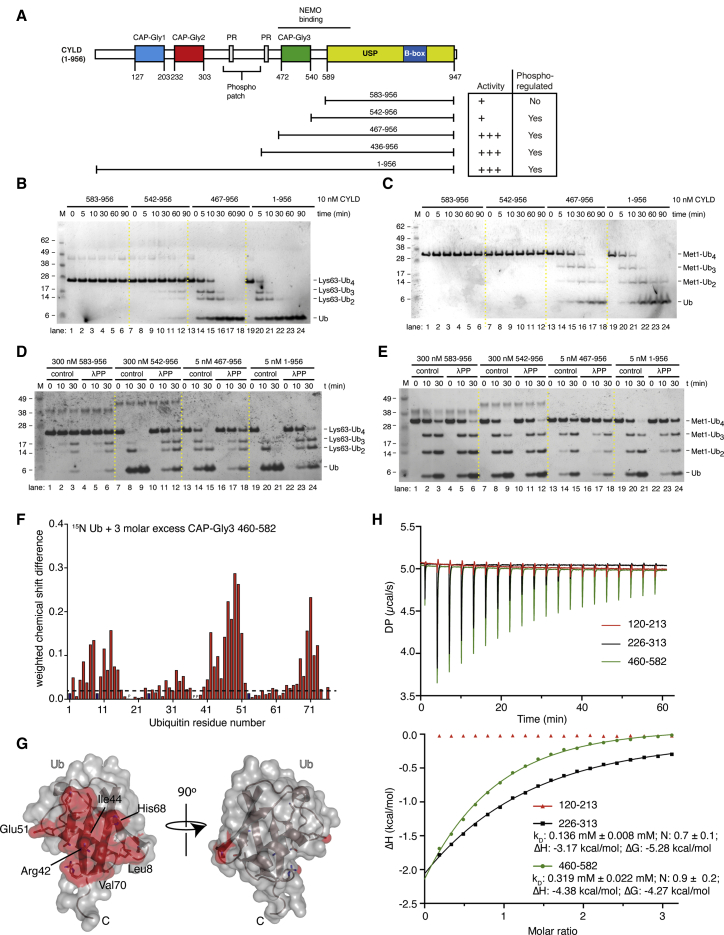
Figure 1.
CYLD is a Lys63-DUB reliant on extra-catalytic domains and phosphorylation for full activity
(A) Schematic representation of the constructs of CYLD generated to test enzymatic activity and reliance on phosphorylation. Table summarizes data shown in subsequent panels.
(B) Qualitative DUB assays for assessing CYLD enzyme activity. Lys63-Ub4 was incubated with different CYLD fragments, and Ub cleavage was monitored over 90 min. Samples were resolved by SDS-PAGE and stained using silver stain.
(C) Qualitative DUB assays as in (B) but using Met1-Ub4 as a substrate. CYLD enzyme activity at 200 nM concentration is shown in Figures S1A and S1B.
(D) Qualitative DUB assay to investigate the phosphorylation status of purified CYLD variants from sf9 cells. sf9-purified CYLD variants were either incubated with λPP or in λPP buffer (control) and then used in a DUB assay as in (B).
(E) As in (D) using Met1-Ub4 chains as substrate.
(F) Chemical shift perturbation plot of Ub for titration of 3-fold excess of CAP-Gly3 (aa 460–582).
(G) Surface of Ub (gray) with chemical shifts mapped from (F) showing that CAP-Gly3 binds the Ub Ile44 patch.
(H) Isothermal titration calorimetry thermograms of the three CYLD CAP-Gly domains. Raw isotherms are shown (top) with integrated fits (bottom) for CAP-Gly2 and CAP-Gly3.